Mdm2 targets the p53 transcription cofactor JMY for degradation
- PMID: 17170761
- PMCID: PMC1796743
- DOI: 10.1038/sj.embor.7400855
Mdm2 targets the p53 transcription cofactor JMY for degradation
Abstract
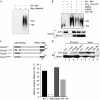
We define here a new mechanism through which Mdm2 (mouse double minute 2) regulates p53 activity, by targeting the p53 transcription cofactor JMY. DNA damage causes an increase in JMY protein, and, in a similar manner, small molecule inhibitors of Mdm2 activity induce JMY in unperturbed cells. At a mechanistic level, Mdm2 regulation of JMY requires the Mdm2 RING (really interesting new gene) finger, which promotes the ubiquitin-dependent degradation of JMY. However, regulation of JMY occurs independently of the p53-binding domain in Mdm2 and p53 activity. These results define a new functional relationship between the p53 cofactor JMY and Mdm2, and indicate that transcription cofactors that facilitate p53 activity are important targets for Mdm2 in suppressing the p53 response.
Figures
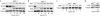



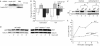
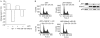
References
-
- Alarcon-Vargas D, Ronai Z (2002) p53–Mdm2—the affair that never ends. Carcinogenesis 23: 541–547 - PubMed
-
- Ardley HC, Robinson PA (2005) E3 ubiquitin ligases. Essays Biochem 41: 15–30 - PubMed
-
- Boyd SD, Tsai KY, Jacks T (2000) An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2: 563–568 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

