Hydrophobic, hydrophilic, and charged amino acid networks within protein
- PMID: 17172302
- PMCID: PMC1914426
- DOI: 10.1529/biophysj.106.098004
Hydrophobic, hydrophilic, and charged amino acid networks within protein
Erratum in
- Biophys J. 2008 Jun;94(11):5081
Abstract
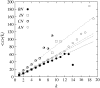
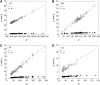
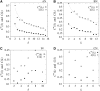
The native three-dimensional structure of a single protein is determined by the physicochemical nature of its constituent amino acids. The 20 different types of amino acids, depending on their physicochemical properties, can be grouped into three major classes: hydrophobic, hydrophilic, and charged. The anatomy of the weighted and unweighted networks of hydrophobic, hydrophilic, and charged residues separately for a large number of proteins were studied. Results showed that the average degree of the hydrophobic networks has a significantly larger value than that of hydrophilic and charged networks. The average degree of the hydrophilic networks is slightly higher than that of the charged networks. The average strength of the nodes of hydrophobic networks is nearly equal to that of the charged network, whereas that of hydrophilic networks has a smaller value than that of hydrophobic and charged networks. The average strength for each of the three types of networks varies with its degree. The average strength of a node in a charged network increases more sharply than that of the hydrophobic and hydrophilic networks. Each of the three types of networks exhibits the "small-world" property. Our results further indicate that the all-amino-acids networks and hydrophobic networks are of assortative type. Although most of the hydrophilic and charged networks are of the assortative type, few others have the characteristics of disassortative mixing of the nodes. We have further observed that all-amino-acids networks and hydrophobic networks bear the signature of hierarchy, whereas the hydrophilic and charged networks do not have any hierarchical signature.
Figures
References
-
- Barabasi, A. L., and R. Albert. 1999. Emergence of scaling in random networks. Science. 286:509–512. - PubMed
-
- Montoya, J. M., and R. V. Sole. 2002. Small world patterns in food webs. J. Theor. Biol. 214:405–412. - PubMed
-
- Watts, D. J., and S. H. Strogatz. 1998. Collective dynamics of ‘small-world’ networks. Nature. 393:440–442. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

