Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor
- PMID: 17172337
- PMCID: PMC1855710
- DOI: 10.1128/JB.01442-06
Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor
Abstract
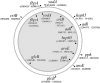
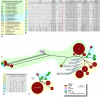
It had been assumed that production of the cytotoxic polyketide mycolactone was strictly associated with Mycobacterium ulcerans, the causative agent of Buruli ulcer. However, a recent study has uncovered a broader distribution of mycolactone-producing mycobacteria (MPM) that includes mycobacteria cultured from diseased fish and frogs in the United States and from diseased fish in the Red and Mediterranean Seas. All of these mycobacteria contain versions of the M. ulcerans pMUM plasmid, produce mycolactones, and show a high degree of genetic relatedness to both M. ulcerans and Mycobacterium marinum. Here, we show by multiple genetic methods, including multilocus sequence analysis and DNA-DNA hybridization, that all MPM have evolved from a common M. marinum progenitor to form a genetically cohesive group among a more diverse assemblage of M. marinum strains. Like M. ulcerans, the fish and frog MPM show multiple copies of the insertion sequence IS2404. Comparisons of pMUM and chromosomal gene sequences demonstrate that plasmid acquisition and the subsequent ability to produce mycolactone were probably the key drivers of speciation. Ongoing evolution among MPM has since produced at least two genetically distinct ecotypes that can be broadly divided into those typically causing disease in ectotherms (but also having a high zoonotic potential) and those causing disease in endotherms, such as humans.
Figures



Similar articles
-
On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer.BMC Genomics. 2012 Jun 19;13:258. doi: 10.1186/1471-2164-13-258. BMC Genomics. 2012. PMID: 22712622 Free PMC article.
-
Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F.Infect Immun. 2006 Nov;74(11):6037-45. doi: 10.1128/IAI.00970-06. Epub 2006 Aug 21. Infect Immun. 2006. PMID: 16923788 Free PMC article.
-
Transfer, stable maintenance and expression of the mycolactone polyketide megasynthase mls genes in a recombination-impaired Mycobacterium marinum.Microbiology (Reading). 2009 Jun;155(Pt 6):1923-1933. doi: 10.1099/mic.0.027029-0. Epub 2009 Apr 21. Microbiology (Reading). 2009. PMID: 19383681
-
The genome, evolution and diversity of Mycobacterium ulcerans.Infect Genet Evol. 2012 Apr;12(3):522-9. doi: 10.1016/j.meegid.2012.01.018. Epub 2012 Jan 28. Infect Genet Evol. 2012. PMID: 22306192 Review.
-
Buruli ulcer and mycolactone-producing mycobacteria.Jpn J Infect Dis. 2013;66(2):83-8. doi: 10.7883/yoken.66.83. Jpn J Infect Dis. 2013. PMID: 23514902 Review.
Cited by
-
Spatiotemporal Co-existence of Two Mycobacterium ulcerans Clonal Complexes in the Offin River Valley of Ghana.PLoS Negl Trop Dis. 2016 Jul 19;10(7):e0004856. doi: 10.1371/journal.pntd.0004856. eCollection 2016 Jul. PLoS Negl Trop Dis. 2016. PMID: 27434064 Free PMC article.
-
On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer.BMC Genomics. 2012 Jun 19;13:258. doi: 10.1186/1471-2164-13-258. BMC Genomics. 2012. PMID: 22712622 Free PMC article.
-
Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar Liflandii.J Bacteriol. 2013 Feb;195(3):556-64. doi: 10.1128/JB.02132-12. Epub 2012 Nov 30. J Bacteriol. 2013. PMID: 23204453 Free PMC article.
-
Could Mycolactone Inspire New Potent Analgesics? Perspectives and Pitfalls.Toxins (Basel). 2019 Sep 4;11(9):516. doi: 10.3390/toxins11090516. Toxins (Basel). 2019. PMID: 31487908 Free PMC article. Review.
-
Aquatic invertebrates as unlikely vectors of Buruli ulcer disease.Emerg Infect Dis. 2008 Aug;14(8):1247-54. doi: 10.3201/eid1408.071503. Emerg Infect Dis. 2008. PMID: 18680648 Free PMC article.
References
-
- Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. - PubMed
-
- Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

