Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay
- PMID: 17172380
- PMCID: PMC2796855
Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay
Abstract
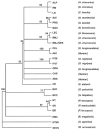
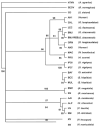
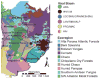
Recently, we reported the discovery of several potential rodent reservoirs of hantaviruses in western (Holochilus chacarius) and eastern Paraguay (Akodon montensis, Oligoryzomys chacoensis, and O. nigripes). Comparisons of the hantavirus S- and M-segments amplified from these four rodents revealed significant differences from each another and from other South American hantaviruses. The ALP strain from the semiarid Chaco ecoregion clustered with Leguna Negra and Rio Mamore (LN/RM), whereas the BMJ-NEB strain from the more humid lower Chaco ecoregion formed a clade with Oran and Bermejo. The other two strains, AAI and IP37/38, were distinct from known hantaviruses. With respect to the S-segment sequence, AAI from eastern Paraguay formed a clade with ALP/LN/RM, but its M-segment clustered with Pergamino and Maciel, suggesting a possible reassortment. AAI was found in areas experiencing rapid land cover fragmentation and change within the Interior Atlantic Forest. IP37/38 did not show any strong association with any of the known hantavirus strains.
Figures
Similar articles
-
Phylogenetic exploration of hantaviruses in Paraguay reveals reassortment and host switching in South America.Virol J. 2011 Aug 12;8:399. doi: 10.1186/1743-422X-8-399. Virol J. 2011. PMID: 21838900 Free PMC article.
-
Laguna Negra virus associated with HPS in western Paraguay and Bolivia.Virology. 1997 Nov 10;238(1):115-27. doi: 10.1006/viro.1997.8840. Virology. 1997. PMID: 9375015
-
Habitat, species richness and hantaviruses of sigmodontine rodents within the Interior Atlantic Forest, Paraguay.PLoS One. 2018 Aug 1;13(8):e0201307. doi: 10.1371/journal.pone.0201307. eCollection 2018. PLoS One. 2018. PMID: 30067840 Free PMC article.
-
Hantavirus reservoirs: current status with an emphasis on data from Brazil.Viruses. 2014 Apr 29;6(5):1929-73. doi: 10.3390/v6051929. Viruses. 2014. PMID: 24784571 Free PMC article. Review.
-
Hantaviruses and cardiopulmonary syndrome in South America.Virus Res. 2014 Jul 17;187:43-54. doi: 10.1016/j.virusres.2014.01.015. Epub 2014 Feb 5. Virus Res. 2014. PMID: 24508343 Review.
Cited by
-
Orthohantavirus diversity in Central-East Argentina: Insights from complete genomic sequencing on phylogenetics, Geographic patterns and transmission scenarios.PLoS Negl Trop Dis. 2024 Oct 9;18(10):e0012465. doi: 10.1371/journal.pntd.0012465. eCollection 2024 Oct. PLoS Negl Trop Dis. 2024. PMID: 39383182 Free PMC article.
-
Impact of Predator Exclusion and Habitat on Seroprevalence of New World Orthohantavirus Harbored by Two Sympatric Rodents within the Interior Atlantic Forest.Viruses. 2021 Sep 29;13(10):1963. doi: 10.3390/v13101963. Viruses. 2021. PMID: 34696393 Free PMC article.
-
Ecology, genetic diversity, and phylogeographic structure of andes virus in humans and rodents in Chile.J Virol. 2009 Mar;83(6):2446-59. doi: 10.1128/JVI.01057-08. Epub 2008 Dec 30. J Virol. 2009. PMID: 19116256 Free PMC article.
-
Immunological mechanisms mediating hantavirus persistence in rodent reservoirs.PLoS Pathog. 2008 Nov;4(11):e1000172. doi: 10.1371/journal.ppat.1000172. Epub 2008 Nov 28. PLoS Pathog. 2008. PMID: 19043585 Free PMC article. Review.
-
Reconstructing the evolutionary origins and phylogeography of hantaviruses.Trends Microbiol. 2014 Aug;22(8):473-82. doi: 10.1016/j.tim.2014.04.008. Epub 2014 May 19. Trends Microbiol. 2014. PMID: 24852723 Free PMC article.
References
-
- Hjelle B, Yates T. Modeling hantavirus maintenance and transmission in rodent communities. Curr Top Microbiol Immunol. 2001;256:77–90. - PubMed
-
- Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. 2001;256:47–75. - PubMed
-
- Plyusnin A. Genetics of hantaviruses: Implications to taxonomy. Arch Virol. 2002;147:665–682. - PubMed
-
- Hinson ER, Shone SM, Zink MC, Glass GE, Klein SL. Wounding: The primary mode of Seoul virus transmission among male Norway rats. Am J Trop Med Hyg. 2004;70:310–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
