Topological dynamics of holins in programmed bacterial lysis
- PMID: 17172454
- PMCID: PMC1750887
- DOI: 10.1073/pnas.0600943103
Topological dynamics of holins in programmed bacterial lysis
Abstract
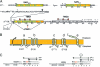
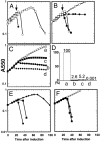
The fate of phage-infected bacteria is determined by the holin, a small membrane protein that triggers to disrupt the membrane at a programmed time, allowing a lysozyme to attack the cell wall. S(21)68, the holin of phage 21, has two transmembrane domains (TMDs) with a predicted N-in, C-in topology. Surprisingly, TMD1 of S(21)68 was found to be dispensable for function, to behave as a SAR ("signal-anchor-release") domain in exiting the membrane to the periplasm, and to engage in homotypic interactions in the soluble phase. The departure of TMD1 from the bilayer coincides with the lethal triggering of the holin and is accelerated by membrane depolarization. Basic residues added at the N terminus of S(21)68 prevent the escape of TMD1 to the periplasm and block hole formation by TMD2. Lysis thus depends on dynamic topology, in that removal of the inhibitory TMD1 from the bilayer frees TMD2 for programmed formation of lethal membrane lesions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
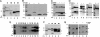
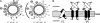
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

