Eg5 steps it up!
- PMID: 17173688
- PMCID: PMC1716758
- DOI: 10.1186/1747-1028-1-31
Eg5 steps it up!
Abstract
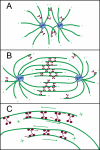
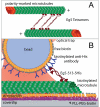
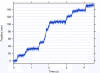
Understanding how molecular motors generate force and move microtubules in mitosis is essential to understanding the physical mechanism of cell division. Recent measurements have shown that one mitotic kinesin superfamily member, Eg5, is mechanically processive and capable of crosslinking and sliding microtubules in vitro. In this review, we highlight recent work that explores how Eg5 functions under load, with an emphasis on the nanomechanical properties of single enzymes.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

