Stabilizing IkappaBalpha by "consensus" design
- PMID: 17174335
- PMCID: PMC1866275
- DOI: 10.1016/j.jmb.2006.11.044
Stabilizing IkappaBalpha by "consensus" design
Abstract
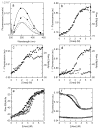
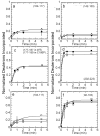
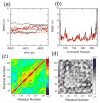
IkappaBalpha is the major regulator of transcription factor NF-kappaB function. The ankyrin repeat region of IkappaBalpha mediates specific interactions with NF-kappaB dimers, but ankyrin repeats 1, 5 and 6 display a highly dynamic character when not in complex with NF-kappaB. Using chemical denaturation, we show here that IkappaBalpha displays two folding transitions: a non-cooperative conversion under weak perturbation, and a major cooperative folding phase upon stronger insult. Taking advantage of a native Trp residue in ankyrin repeat (AR) 6 and engineered Trp residues in AR2, AR4 and AR5, we show that the cooperative transition involves AR2 and AR3, while the non-cooperative transition involves AR5 and AR6. The major structural transition can be affected by single amino acid substitutions converging to the "consensus" ankyrin repeat sequence, increasing the native state stability significantly. We further characterized the structural and dynamic properties of the native state ensemble of IkappaBalpha and the stabilized mutants by H/(2)H exchange mass spectrometry and NMR. The solution experiments were complemented with molecular dynamics simulations to elucidate the microscopic origins of the stabilizing effect of the consensus substitutions, which can be traced to the fast conformational dynamics of the folded ensemble.
Figures

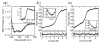


References
-
- Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. - PubMed
-
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Ann Rev Immunol. 1996;14:649–83. - PubMed
-
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Ann Rev Immunol. 1998;16:225–60. - PubMed
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

