Identification of 3-deoxyglucosone dehydrogenase as aldehyde dehydrogenase 1A1 (retinaldehyde dehydrogenase 1)
- PMID: 17175089
- PMCID: PMC1885224
- DOI: 10.1016/j.biochi.2006.11.005
Identification of 3-deoxyglucosone dehydrogenase as aldehyde dehydrogenase 1A1 (retinaldehyde dehydrogenase 1)
Abstract
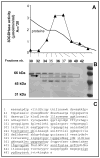
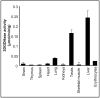
One of the metabolic fates of 3-deoxyglucosone, a product of protein deglycation and a potent glycating agent, is to be oxidized to 2-keto-3-deoxygluconate, but the enzyme that catalyzes this reaction is presently unknown. Starting from human erythrocytes, which are known to convert 3-deoxyglucosone to 2-keto-3-deoxygluconate, we have purified to near homogeneity a NAD-dependent dehydrogenase that catalyzes this last reaction at neutral pH. Sequencing of a 55 kDa band co-eluting with the enzymatic activity in the last step indicated that it corresponded to aldehyde dehydrogenase 1A1 (ALDH1A1), an enzyme known to catalyze the oxidation of retinaldehyde to retinoic acid. Overexpression of human ALDH1A1 in HEK cells led to a more than 20-fold increase in 3-deoxyglucosone dehydrogenase activity. In mouse tissues 3-deoxyglucosone dehydrogenase activity was highest in liver, intermediate in lung and testis, and negligible or undetectable in other tissues, in agreement with the tissue distribution of ALDH1A1 mRNA. 3-deoxyglucosone dehydrogenase activity was undetectable in tissues from ALDH1A1(-/-) mice. ALDH1A1 appears therefore to be the major if not the only enzyme responsible for the oxidation of 3-deoxyglucosone to 2-keto-3-deoxygluconate. The urinary excretion of 2-keto-3-deoxygluconate amounted to 16.7 micromol/g creatinine in humans, indicating that 3-deoxyglucosone may be quantitatively a more important substrate than retinaldehyde for ALDH1A1.
Figures
Similar articles
-
Enzymatic conversion of retinaldehyde to retinoic acid by cloned murine cytosolic and mitochondrial aldehyde dehydrogenases.Mol Pharmacol. 1994 Jul;46(1):88-96. Mol Pharmacol. 1994. PMID: 8058062
-
Structural and kinetic features of aldehyde dehydrogenase 1A (ALDH1A) subfamily members, cancer stem cell markers active in retinoic acid biosynthesis.Arch Biochem Biophys. 2020 Mar 15;681:108256. doi: 10.1016/j.abb.2020.108256. Epub 2020 Jan 7. Arch Biochem Biophys. 2020. PMID: 31923393
-
Cloning of a rat cDNA encoding retinal dehydrogenase isozyme type I and its expression in E. coli.Gene. 1997 Jun 3;191(2):167-72. doi: 10.1016/s0378-1119(97)00054-1. Gene. 1997. PMID: 9218716
-
Aldehyde dehydrogenase 1A1: friend or foe to female metabolism?Nutrients. 2014 Mar 3;6(3):950-73. doi: 10.3390/nu6030950. Nutrients. 2014. PMID: 24594504 Free PMC article. Review.
-
Aldehyde dehydrogenase 1A1 and 1A3 isoforms - mechanism of activation and regulation in cancer.Cell Signal. 2021 Nov;87:110120. doi: 10.1016/j.cellsig.2021.110120. Epub 2021 Aug 21. Cell Signal. 2021. PMID: 34428540 Free PMC article. Review.
Cited by
-
Glyoxalase System as a Therapeutic Target against Diabetic Retinopathy.Antioxidants (Basel). 2020 Oct 30;9(11):1062. doi: 10.3390/antiox9111062. Antioxidants (Basel). 2020. PMID: 33143048 Free PMC article. Review.
-
The contribution of vitamin A to autocrine regulation of fat depots.Biochim Biophys Acta. 2012 Jan;1821(1):190-7. doi: 10.1016/j.bbalip.2011.06.004. Epub 2011 Jun 13. Biochim Biophys Acta. 2012. PMID: 21704731 Free PMC article. Review.
-
Dicarbonyls and glyoxalase in disease mechanisms and clinical therapeutics.Glycoconj J. 2016 Aug;33(4):513-25. doi: 10.1007/s10719-016-9705-z. Epub 2016 Jul 12. Glycoconj J. 2016. PMID: 27406712 Free PMC article. Review.
-
Differential gene expression analysis of subcutaneous fat, fascia, and skin overlying a Dupuytren's disease nodule in comparison to control tissue.Hand (N Y). 2009 Sep;4(3):294-301. doi: 10.1007/s11552-009-9164-0. Epub 2009 Jan 29. Hand (N Y). 2009. PMID: 19184239 Free PMC article.
-
Concerted action of aldehyde dehydrogenases influences depot-specific fat formation.Mol Endocrinol. 2011 May;25(5):799-809. doi: 10.1210/me.2010-0465. Epub 2011 Mar 24. Mol Endocrinol. 2011. PMID: 21436255 Free PMC article.
References
-
- Delpierre G, Rider MH, Collard F, Stroobant V, Vanstapel F, Santos H, Van Schaftingen E. Identification, cloning, and heterologous expression of a mammalian fructosamine-3-kinase. Diabetes. 2000;49:1627–1634. - PubMed
-
- Szwergold BS, Howell S, Beisswenger PJ. Human fructosamine-3-kinase: purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes. 2001;50:2139–2147. - PubMed
-
- Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. The Amadori product on protein: structure and reactions. Prog Clin Biol Res. 1989;304:43–67. - PubMed
-
- Niwa T. 3-Deoxyglucosone: metabolism, analysis, biological activity, and clinical implication. J Chromatogr B Biomed Sci Appl. 1999;731:23–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

