Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3'- and/or 5'-ssDNA branches
- PMID: 17176115
- PMCID: PMC2528077
- DOI: 10.1021/bi061626q
Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3'- and/or 5'-ssDNA branches
Abstract
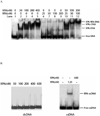
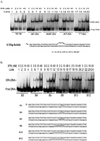
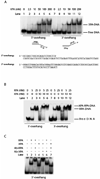
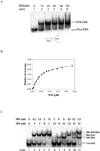
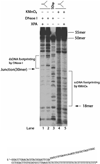
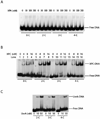
Human XPA is an important DNA damage recognition protein in nucleotide excision repair (NER). We previously observed that XPA binds to the DNA lesion as a homodimer [Liu, Y., Liu, Y., Yang, Z., Utzat, C., Wang, G., Basu, A. K., and Zou, Y. (2005) Biochemistry 44, 7361-7368]. Herein we report that XPA recognized undamaged DNA double-strand/single-strand (ds-ssDNA) junctions containing ssDNA branches with binding affinity (Kd = 49.1 +/- 5.1 nM) much higher than its ability to bind to DNA damage. The recognized DNA junction structures include the Y-shape junction (with both 3'- and 5'-ssDNA branches), 3'-overhang junction (with a 3'-ssDNA branch), and 5'-overhang junction (with a 5'-ssDNA branch). Using gel filtration chromatography and gel mobility shift assays, we showed that the highly efficient binding appeared to be carried out by the XPA monomer and that the binding was largely independent of RPA. Furthermore, XPA efficiently bound to six-nucleotide mismatched DNA bubble substrates with or without DNA adducts including C8 guanine adducts of AF, AAF, and AP and the T[6,4]T photoproducts. Using a set of defined DNA substrates with varying degrees of DNA bending, we also found that the XPC-HR23B complex recognized DNA bending, whereas neither XPA nor the XPA-RPA complex could bind to bent DNA. We propose that, besides DNA damage recognition, XPA may also play a novel role in stabilizing, via its high affinity to ds-ssDNA junctions, the DNA strand opening surrounding the lesion for stable formation of preincision NER intermediates. Our results provide a plausible mechanistic interpretation for the indispensable requirement of XPA for both global genome and transcription-coupled repairs. Since ds-ssDNA junctions are common intermediates in many DNA metabolic pathways, the additional potential role of XPA in cellular processes is discussed.
Figures

References
-
- Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. - PubMed
-
- Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog. 2003;38:1–13. - PubMed
-
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. - PubMed
-
- Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

