Insights on the evolution of trehalose biosynthesis
- PMID: 17178000
- PMCID: PMC1769515
- DOI: 10.1186/1471-2148-6-109
Insights on the evolution of trehalose biosynthesis
Abstract
Background: The compatible solute trehalose is a non-reducing disaccharide, which accumulates upon heat, cold or osmotic stress. It was commonly accepted that trehalose is only present in extremophiles or cryptobiotic organisms. However, in recent years it has been shown that although higher plants do not accumulate trehalose at significant levels they have actively transcribed genes encoding the corresponding biosynthetic enzymes.
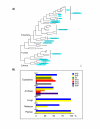
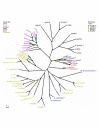
Results: In this study we show that trehalose biosynthesis ability is present in eubacteria, archaea, plants, fungi and animals. In bacteria there are five different biosynthetic routes, whereas in fungi, plants and animals there is only one. We present phylogenetic analyses of the trehalose-6-phosphate synthase (TPS) and trehalose-phosphatase (TPP) domains and show that there is a close evolutionary relationship between these domains in proteins from diverse organisms. In bacteria TPS and TPP genes are clustered, whereas in eukaryotes these domains are fused in a single protein.
Conclusion: We have demonstrated that trehalose biosynthesis pathways are widely distributed in nature. Interestingly, several eubacterial species have multiple pathways, while eukaryotes have only the TPS/TPP pathway. Vertebrates lack trehalose biosynthetic capacity but can catabolise it. TPS and TPP domains have evolved mainly in parallel and it is likely that they have experienced several instances of gene duplication and lateral gene transfer.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

