Encoding folding paths of RNA switches
- PMID: 17178750
- PMCID: PMC1802593
- DOI: 10.1093/nar/gkl1036
Encoding folding paths of RNA switches
Abstract
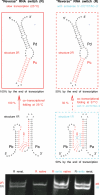
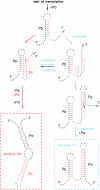
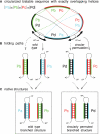
RNA co-transcriptional folding has long been suspected to play an active role in helping proper native folding of ribozymes and structured regulatory motifs in mRNA untranslated regions (UTRs). Yet, the underlying mechanisms and coding requirements for efficient co-transcriptional folding remain unclear. Traditional approaches have intrinsic limitations to dissect RNA folding paths, as they rely on sequence mutations or circular permutations that typically perturb both RNA folding paths and equilibrium structures. Here, we show that exploiting sequence symmetries instead of mutations can circumvent this problem by essentially decoupling folding paths from equilibrium structures of designed RNA sequences. Using bistable RNA switches with symmetrical helices conserved under sequence reversal, we demonstrate experimentally that native and transiently formed helices can guide efficient co-transcriptional folding into either long-lived structure of these RNA switches. Their folding path is controlled by the order of helix nucleations and subsequent exchanges during transcription, and may also be redirected by transient antisense interactions. Hence, transient intra- and inter-molecular base pair interactions can effectively regulate the folding of nascent RNA molecules into different native structures, provided limited coding requirements, as discussed from an information theory perspective. This constitutive coupling between RNA synthesis and RNA folding regulation may have enabled the early emergence of autonomous RNA-based regulation networks.
Figures



References
-
- Dahlberg A.E. The ribosome in action. Science. 2001;292:868–869. - PubMed
-
- Kruger K., Grabowski P., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. - PubMed
-
- Bartel D.P., Szostak J.W. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. - PubMed
-
- Joyce G.F. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82:83–87. - PubMed
-
- Ellington A.E., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed

