High resolution array-CGH analysis of single cells
- PMID: 17178751
- PMCID: PMC1807964
- DOI: 10.1093/nar/gkl1030
High resolution array-CGH analysis of single cells
Abstract
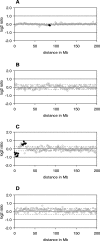
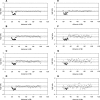
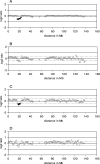
Heterogeneity in the genome copy number of tissues is of particular importance in solid tumor biology. Furthermore, many clinical applications such as pre-implantation and non-invasive prenatal diagnosis would benefit from the ability to characterize individual single cells. As the amount of DNA from single cells is so small, several PCR protocols have been developed in an attempt to achieve unbiased amplification. Many of these approaches are suitable for subsequent cytogenetic analyses using conventional methodologies such as comparative genomic hybridization (CGH) to metaphase spreads. However, attempts to harness array-CGH for single-cell analysis to provide improved resolution have been disappointing. Here we describe a strategy that combines single-cell amplification using GenomePlex library technology (GenomePlex) Single Cell Whole Genome Amplification Kit, Sigma-Aldrich, UK) and detailed analysis of genomic copy number changes by high-resolution array-CGH. We show that single copy changes as small as 8.3 Mb in single cells are detected reliably with single cells derived from various tumor cell lines as well as patients presenting with trisomy 21 and Prader-Willi syndrome. Our results demonstrate the potential of this technology for studies of tumor biology and for clinical diagnostics.
Figures




References
-
- Telenius H., Carter N.P., Bebb C.E., Nordenskjold M., Ponder B.A., Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. - PubMed
-
- Speicher M.R., du Manoir S., Schrock E., Holtgreve-Grez H., Schoell B., Lengauer C., Cremer T., Ried T. Molecular cytogenetic analysis of formalin-fixed, paraffin-embedded solid tumors by comparative genomic hybridization after universal DNA-amplification. Hum. Mol. Genet. 1993;2:1907–1914. - PubMed
-
- Speicher M.R., Howe C., Crotty P., du Manoir S., Costa J., Ward D.C. Comparative genomic hybridization detects novel deletions and amplifications in head and neck squamous cell carcinomas. Cancer Res. 1995;55:1010–1013. - PubMed
-
- Speicher M.R., Jauch A., Walt H., du Manoir S., Ried T., Jochum W., Sulser T., Cremer T. Correlation of microscopic phenotype with genotype in a formalin-fixed, paraffin-embedded testicular germ cell tumor with universal DNA amplification, comparative genomic hybridization, and interphase cytogenetics. Am. J. Pathol. 1995;146:1332–1340. - PMC - PubMed
-
- Wiltshire R.N., Duray P., Bittner M.L., Visakorpi T., Meltzer P.S., Tuthill R.J., Liotta L.A., Trent J.M. Direct visualization of the clonal progression of primary cutaneous melanoma: application of tissue microdissection and comparative genomic hybridization. Cancer Res. 1995;55:3954–3957. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

