Protease gene families in Populus and Arabidopsis
- PMID: 17181860
- PMCID: PMC1780054
- DOI: 10.1186/1471-2229-6-30
Protease gene families in Populus and Arabidopsis
Abstract
Background: Proteases play key roles in plants, maintaining strict protein quality control and degrading specific sets of proteins in response to diverse environmental and developmental stimuli. Similarities and differences between the proteases expressed in different species may give valuable insights into their physiological roles and evolution.
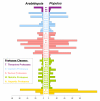
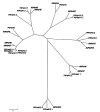
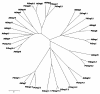
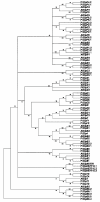
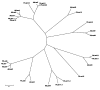
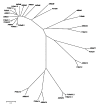
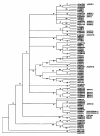
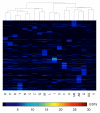
Results: We have performed a comparative analysis of protease genes in the two sequenced dicot genomes, Arabidopsis thaliana and Populus trichocarpa by using genes coding for proteases in the MEROPS database 1 for Arabidopsis to identify homologous sequences in Populus. A multigene-based phylogenetic analysis was performed. Most protease families were found to be larger in Populus than in Arabidopsis, reflecting recent genome duplication. Detailed studies on e.g. the DegP, Clp, FtsH, Lon, rhomboid and papain-Like protease families showed the pattern of gene family expansion and gene loss was complex. We finally show that different Populus tissues express unique suites of protease genes and that the mRNA levels of different classes of proteases change along a developmental gradient.
Conclusion: Recent gene family expansion and contractions have made the Arabidopsis and Populus complements of proteases different and this, together with expression patterns, gives indications about the roles of the individual gene products or groups of proteases.
Figures

Similar articles
-
Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar.J Integr Plant Biol. 2010 Nov;52(11):1016-26. doi: 10.1111/j.1744-7909.2010.00982.x. J Integr Plant Biol. 2010. PMID: 20977659
-
Genome-wide analysis of the MADS-box gene family in Populus trichocarpa.Gene. 2006 Aug 15;378:84-94. doi: 10.1016/j.gene.2006.05.022. Epub 2006 Jul 10. Gene. 2006. PMID: 16831523
-
Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa.Biochem Biophys Res Commun. 2008 Jul 4;371(3):468-74. doi: 10.1016/j.bbrc.2008.04.087. Epub 2008 Apr 28. Biochem Biophys Res Commun. 2008. PMID: 18442469
-
Genome-wide analysis of plant glutaredoxin systems.J Exp Bot. 2006;57(8):1685-96. doi: 10.1093/jxb/erl001. Epub 2006 May 23. J Exp Bot. 2006. PMID: 16720602 Review.
-
The endo-beta-mannanase gene families in Arabidopsis, rice, and poplar.Funct Integr Genomics. 2007 Jan;7(1):1-16. doi: 10.1007/s10142-006-0034-3. Epub 2006 Aug 8. Funct Integr Genomics. 2007. PMID: 16897088 Review.
Cited by
-
The poplar-poplar rust interaction: insights from genomics and transcriptomics.J Pathog. 2011;2011:716041. doi: 10.4061/2011/716041. Epub 2011 Oct 26. J Pathog. 2011. PMID: 22567338 Free PMC article.
-
The SUMO conjugation pathway in Populus: genomic analysis, tissue-specific and inducible SUMOylation and in vitro de-SUMOylation.Planta. 2010 Jun;232(1):51-9. doi: 10.1007/s00425-010-1151-8. Epub 2010 Apr 2. Planta. 2010. PMID: 20361336
-
A new principle of oligomerization of plant DEG7 protease based on interactions of degenerated protease domains.Biochem J. 2011 Apr 1;435(1):167-74. doi: 10.1042/BJ20101613. Biochem J. 2011. PMID: 21247409 Free PMC article.
-
Programmed cell death in plants: A chloroplastic connection.Plant Signal Behav. 2015;10(2):e989752. doi: 10.4161/15592324.2014.989752. Plant Signal Behav. 2015. PMID: 25760871 Free PMC article. Review.
-
Sequence comparison, molecular modeling, and network analysis predict structural diversity in cysteine proteases from the Cape sundew, Drosera capensis.Comput Struct Biotechnol J. 2016 Jun 14;14:271-82. doi: 10.1016/j.csbj.2016.05.003. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27471585 Free PMC article.
References
-
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–604. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

