Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry
- PMID: 17182681
- PMCID: PMC1865946
- DOI: 10.1128/JVI.02006-06
Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry
Abstract
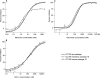
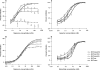
Maraviroc is a CCR5 antagonist in clinical development as one of a new class of antiretrovirals targeting human immunodeficiency virus type 1 (HIV-1) coreceptor binding. We investigated the mechanism of HIV resistance to maraviroc by using in vitro sequential passage and site-directed mutagenesis. Serial passage through increasing maraviroc concentrations failed to select maraviroc-resistant variants from some laboratory-adapted and clinical isolates of HIV-1. However, high-level resistance to maraviroc was selected from three of six primary isolates passaged in peripheral blood lymphocytes (PBL). The SF162 strain acquired resistance to maraviroc in both treated and control cultures; all resistant variants were able to use CXCR4 as a coreceptor. In contrast, maraviroc-resistant virus derived from isolates CC1/85 and RU570 remained CCR5 tropic, as evidenced by susceptibility to the CCR5 antagonist SCH-C, resistance to the CXCR4 antagonist AMD3100, and an inability to replicate in CCR5 Delta32/Delta32 PBL. Strain-specific mutations were identified in the V3 loop of maraviroc-resistant CC1/85 and RU570. The envelope-encoding region of maraviroc-resistant CC1/85 was inserted into an NL4-3 background. This recombinant virus was completely resistant to maraviroc but retained susceptibility to aplaviroc. Reverse mutation of gp120 residues 316 and 323 in the V3 loop (numbering from HXB2) to their original sequence restored wild-type susceptibility to maraviroc, while reversion of either mutation resulted in a partially sensitive virus with reduced maximal inhibition (plateau). The plateaus are consistent with the virus having acquired the ability to utilize maraviroc-bound receptor for entry. This hypothesis was further corroborated by the observation that a high concentration of maraviroc blocks the activity of aplaviroc against maraviroc-resistant virus.
Figures
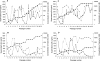
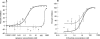
References
-
- Cheng-Mayer, C., and J. A. Levy. 1988. Distinct biological and serological properties of human immunodeficiency viruses from the brain. Ann. Neurol. 23(Suppl.):S58-S61. - PubMed
-
- Christopoulos, A., and T. Kenakin. 2002. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54:323-374. - PubMed
-
- Coakley, E., C. J. Petropoulos, and J. M. Whitcomb. 2005. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 18:9-15. - PubMed
-
- Dejucq, N., G. Simmons, and P. R. Clapham. 2000. T-cell line adaptation of human immunodeficiency virus type 1 strain SF162: effects on envelope, vpu and macrophage-tropism. J. Gen. Virol. 81:2899-2904. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

