Inhibition of filovirus replication by the zinc finger antiviral protein
- PMID: 17182693
- PMCID: PMC1865956
- DOI: 10.1128/JVI.01601-06
Inhibition of filovirus replication by the zinc finger antiviral protein
Abstract
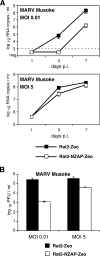
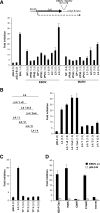
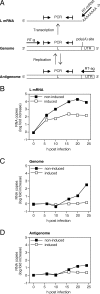
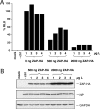
The zinc finger antiviral protein (ZAP) was recently shown to inhibit Moloney murine leukemia virus and Sindbis virus replication. We tested whether ZAP also acts against Ebola virus (EBOV) and Marburg virus (MARV). Antiviral effects were observed after infection of cells expressing the N-terminal part of ZAP fused to the product of the zeocin resistance gene (NZAP-Zeo) as well as after infection of cells inducibly expressing full-length ZAP. EBOV was inhibited by up to 4 log units, whereas MARV was inhibited between 1 to 2 log units. The activity of ZAP was dependent on the integrity of the second and fourth zinc finger motif, as tested with cell lines expressing NZAP-Zeo mutants. Heterologous expression of EBOV- and MARV-specific sequences fused to a reporter gene suggest that ZAP specifically targets L gene sequences. The activity of NZAP-Zeo in this assay was also dependent on the integrity of the second and fourth zinc finger motif. Time-course experiments with infectious EBOV showed that ZAP reduces the level of L mRNA before the level of genomic or antigenomic RNA is affected. Transient expression of ZAP decreased the activity of an EBOV replicon system by up to 95%. This inhibitory effect could be partially compensated for by overexpression of L protein. In conclusion, the data demonstrate that ZAP exhibits antiviral activity against filoviruses, presumably by decreasing the level of viral mRNA.
Figures
References
-
- Boehmann, Y., S. Enterlein, A. Randolf, and E. Mühlberger. 2005. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology 332:406-417. - PubMed
-
- Bowen, E. T., G. Lloyd, W. J. Harris, G. S. Platt, A. Baskerville, and E. E. Vella. 1977. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet i:571-573. - PubMed
-
- Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

