Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells
- PMID: 17182852
- PMCID: PMC1805101
- DOI: 10.1091/mbc.e06-07-0587
Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells
Abstract
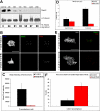
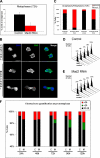
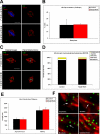
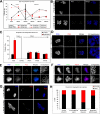
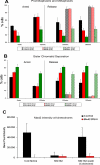
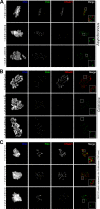
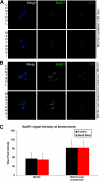
The spindle assembly checkpoint is essential to maintain genomic stability during cell division. We analyzed the role of the putative Drosophila Mad2 homologue in the spindle assembly checkpoint and mitotic progression. Depletion of Mad2 by RNAi from S2 cells shows that it is essential to prevent mitotic exit after spindle damage, demonstrating its conserved role. Mad2-depleted cells also show accelerated transit through prometaphase and premature sister chromatid separation, fail to form metaphases, and exit mitosis soon after nuclear envelope breakdown with extensive chromatin bridges that result in severe aneuploidy. Interestingly, preventing Mad2-depleted cells from exiting mitosis by a checkpoint-independent arrest allows congression of normally condensed chromosomes. More importantly, a transient mitotic arrest is sufficient for Mad2-depleted cells to exit mitosis with normal patterns of chromosome segregation, suggesting that all the associated phenotypes result from a highly accelerated exit from mitosis. Surprisingly, if Mad2-depleted cells are blocked transiently in mitosis and then released into a media containing a microtubule poison, they arrest with high levels of kinetochore-associated BubR1, properly localized cohesin complex and fail to exit mitosis revealing normal spindle assembly checkpoint activity. This behavior is specific for Mad2 because BubR1-depleted cells fail to arrest in mitosis under these experimental conditions. Taken together our results strongly suggest that Mad2 is exclusively required to delay progression through early stages of prometaphase so that cells have time to fully engage the spindle assembly checkpoint, allowing a controlled metaphase-anaphase transition and normal patterns of chromosome segregation.
Figures
Similar articles
-
Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster.Mol Cell Biol. 2010 Jul;30(13):3384-95. doi: 10.1128/MCB.00258-10. Epub 2010 Apr 26. Mol Cell Biol. 2010. PMID: 20421417 Free PMC article.
-
Spindle checkpoint-independent inhibition of mitotic chromosome segregation by Drosophila Mps1.Mol Biol Cell. 2012 Jun;23(12):2275-91. doi: 10.1091/mbc.E12-02-0117. Epub 2012 May 2. Mol Biol Cell. 2012. PMID: 22553353 Free PMC article.
-
BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis.Cell Cycle. 2007 Jun 1;6(11):1367-78. doi: 10.4161/cc.6.11.4271. Epub 2007 Jun 11. Cell Cycle. 2007. PMID: 17525528
-
Dual inhibition of Cdc20 by the spindle checkpoint.J Biomed Sci. 2007 Jul;14(4):475-9. doi: 10.1007/s11373-007-9157-3. Epub 2007 Mar 17. J Biomed Sci. 2007. PMID: 17370142 Review.
-
The Mad1-Mad2 balancing act--a damaged spindle checkpoint in chromosome instability and cancer.J Cell Sci. 2012 Sep 15;125(Pt 18):4197-206. doi: 10.1242/jcs.107037. Epub 2012 Oct 23. J Cell Sci. 2012. PMID: 23093575 Review.
Cited by
-
Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons.Proc Natl Acad Sci U S A. 2012 Aug 14;109(33):E2205-14. doi: 10.1073/pnas.1201911109. Epub 2012 Jul 9. Proc Natl Acad Sci U S A. 2012. PMID: 22778409 Free PMC article.
-
Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster.Mol Cell Biol. 2010 Jul;30(13):3384-95. doi: 10.1128/MCB.00258-10. Epub 2010 Apr 26. Mol Cell Biol. 2010. PMID: 20421417 Free PMC article.
-
A potent estrogen receptor and microtubule specific purine-benzothiazole-based fluorescent molecular probe induces apoptotic death of breast cancer cells.Sci Rep. 2022 Jun 24;12(1):10772. doi: 10.1038/s41598-022-12933-8. Sci Rep. 2022. PMID: 35750870 Free PMC article.
-
Deregulated Aurora-B induced tetraploidy promotes tumorigenesis.FASEB J. 2009 Aug;23(8):2741-8. doi: 10.1096/fj.09-130963. Epub 2009 Mar 30. FASEB J. 2009. PMID: 19332642 Free PMC article.
-
Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis.Mol Biol Cell. 2007 Dec;18(12):4847-58. doi: 10.1091/mbc.e06-06-0539. Epub 2007 Sep 19. Mol Biol Cell. 2007. PMID: 17881737 Free PMC article.
References
-
- Bharadwaj R., Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. - PubMed
-
- Bhat M. A., Philp A. V., Glover D. M., Bellen H. J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
-
- Buffin E., Lefebvre C., Huang J., Gagou M. E., Karess R. E. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 2005;15:856–861. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

