Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation
- PMID: 17182856
- PMCID: PMC1805084
- DOI: 10.1091/mbc.e06-05-0458
Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation
Abstract
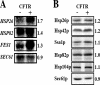
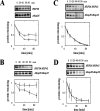
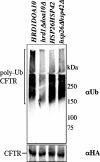
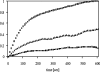
Secreted proteins that fail to achieve their native conformations, such as cystic fibrosis transmembrane conductance regulator (CFTR) and particularly the DeltaF508-CFTR variant can be selected for endoplasmic reticulum (ER)-associated degradation (ERAD) by molecular chaperones. Because the message corresponding to HSP26, which encodes a small heat-shock protein (sHsp) in yeast was up-regulated in response to CFTR expression, we examined the impact of sHsps on ERAD. First, we observed that CFTR was completely stabilized in cells lacking two partially redundant sHsps, Hsp26p and Hsp42p. Interestingly, the ERAD of a soluble and a related integral membrane protein were unaffected in yeast deleted for the genes encoding these sHsps, and CFTR polyubiquitination was also unaltered, suggesting that Hsp26p/Hsp42p are not essential for polyubiquitination. Next, we discovered that DeltaF508-CFTR degradation was enhanced when a mammalian sHsp, alphaA-crystallin, was overexpressed in human embryonic kidney 293 cells, but wild-type CFTR biogenesis was unchanged. Because alphaA-crystallin interacted preferentially with DeltaF508-CFTR and because purified alphaA-crystallin suppressed the aggregation of the first nucleotide-binding domain of CFTR, we suggest that sHsps maintain the solubility of DeltaF508-CFTR during the ERAD of this polypeptide.
Figures




References
-
- Ahner A., Brodsky J. L. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. - PubMed
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidmann J. G., Smith J. G., Struhl K. New York: Greene Publishing Associates and Wiley Interscience; 1988. Current protocols in Molecular Biology.
-
- Basha E., Friedrich K. L., Vierling E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 2006;281:39943–39952. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

