Mitf regulation of Dia1 controls melanoma proliferation and invasiveness
- PMID: 17182868
- PMCID: PMC1698449
- DOI: 10.1101/gad.406406
Mitf regulation of Dia1 controls melanoma proliferation and invasiveness
Abstract
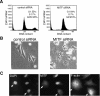
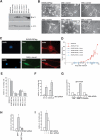
It is widely held that cells with metastatic properties such as invasiveness and expression of matrix metalloproteinases arise through the stepwise accumulation of genetic lesions arising from genetic instability and "clonal evolution." By contrast, we show here that in melanomas invasiveness can be regulated epigenetically by the microphthalmia-associated transcription factor, Mitf, via regulation of the DIAPH1 gene encoding the diaphanous-related formin Dia1 that promotes actin polymerization and coordinates the actin cytoskeleton and microtubule networks at the cell periphery. Low Mitf levels lead to down-regulation of Dia1, reorganization of the actin cytoskeleton, and increased ROCK-dependent invasiveness, whereas increased Mitf expression leads to decreased invasiveness. Significantly the regulation of Dia1 by Mitf also controls p27(Kip1)-degradation such that reduced Mitf levels lead to a p27(Kip1)-dependent G1 arrest. Thus Mitf, via regulation of Dia1, can both inhibit invasiveness and promote proliferation. The results imply variations in the repertoire of environmental cues that determine Mitf activity will dictate the differentiation, proliferative, and invasive/migratory potential of melanoma cells through a dynamic epigenetic mechanism.
Figures





References
-
- Alberts, A.S. Diaphanous-related Formin homology proteins. Curr. Biol. 2002;12:R796. - PubMed
-
- Alonso, S.R., Ortiz, P., Pollan, M., Perez-Gomez, B., Sanchez, L., Acuna, M.J., Pajares, R., Martinez-Tello, F.J., Hortelano, C.M., Piris, M.A., et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: A tissue microarray-based study. Am. J. Pathol. 2004;164:193–203. - PMC - PubMed
-
- Arber, S., Barbayannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., Caroni, P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. - PubMed
-
- Bales, E.S., Dietrich, C., Bandyopadhyay, D., Schwahn, D.J., Xu, W., Didenko, V., Leiss, P., Conrad, N., Pereira-Smith, O., Orengo, I., et al. High levels of expression of p27KIP1 and cyclin E in invasive primary malignant melanomas. J. Invest. Dermatol. 1999;113:1039–1046. - PubMed
-
- Baylin, S.B., Ohm, J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
