High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer
- PMID: 17184525
- PMCID: PMC1769510
- DOI: 10.1186/1471-2407-6-295
High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer
Abstract
Background: The development of targeted therapies has created a pressing clinical need for the rapid and robust molecular characterisation of cancers. We describe here the application of high-resolution melting analysis (HRM) to screen for KRAS mutations in clinical cancer samples. In non-small cell lung cancer, KRAS mutations have been shown to identify a group of patients that do not respond to EGFR targeted therapies and the identification of these mutations is thus clinically important.
Methods: We developed a high-resolution melting (HRM) assay to detect somatic mutations in exon 2, notably codons 12 and 13 of the KRAS gene using the intercalating dye SYTO 9. We tested 3 different cell lines with known KRAS mutations and then examined the sensitivity of mutation detection with the cell lines using 189 bp and 92 bp amplicons spanning codons 12 and 13. We then screened for KRAS mutations in 30 non-small cell lung cancer biopsies that had been previously sequenced for mutations in EGFR exons 18-21.
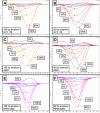
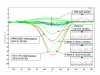
Results: Known KRAS mutations in cell lines (A549, HCT116 and RPMI8226) were readily detectable using HRM. The shorter 92 bp amplicon was more sensitive in detecting mutations than the 189 bp amplicon and was able to reliably detect as little as 5-6% of each cell line DNA diluted in normal DNA. Nine of the 30 non-small cell lung cancer biopsies had KRAS mutations detected by HRM analysis. The results were confirmed by standard sequencing. Mutations in KRAS and EGFR were mutually exclusive.
Conclusion: HRM is a sensitive in-tube methodology to screen for mutations in clinical samples. HRM will enable high-throughput screening of gene mutations to allow appropriate therapeutic choices for patients and accelerate research aimed at identifying novel mutations in human cancer.
Figures


Similar articles
-
High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies.BMC Cancer. 2008 May 21;8:142. doi: 10.1186/1471-2407-8-142. BMC Cancer. 2008. PMID: 18495026 Free PMC article.
-
Detection of EGFR and KRAS mutations on trans-thoracic needle aspiration of lung nodules by high resolution melting analysis.J Clin Pathol. 2009 Dec;62(12):1096-102. doi: 10.1136/jcp.2009.067587. Epub 2009 Jul 28. J Clin Pathol. 2009. PMID: 19640859
-
Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing.BMC Cancer. 2011 Sep 24;11:406. doi: 10.1186/1471-2407-11-406. BMC Cancer. 2011. PMID: 21943394 Free PMC article.
-
KRAS mutations in lung cancer.Clin Lung Cancer. 2013 May;14(3):205-14. doi: 10.1016/j.cllc.2012.09.007. Epub 2012 Nov 1. Clin Lung Cancer. 2013. PMID: 23122493 Review.
-
Selumetinib in advanced non small cell lung cancer (NSCLC) harbouring KRAS mutation: endless clinical challenge to KRAS-mutant NSCLC.Rev Recent Clin Trials. 2013 Jun;8(2):93-100. doi: 10.2174/15748871113089990047. Rev Recent Clin Trials. 2013. PMID: 24063423 Review.
Cited by
-
Frequency of TP53 Mutations and its Impact on Drug Sensitivity in Acute Myeloid Leukemia?Indian J Clin Biochem. 2012 Apr;27(2):121-6. doi: 10.1007/s12291-012-0203-1. Epub 2012 Mar 24. Indian J Clin Biochem. 2012. Retraction in: Indian J Clin Biochem. 2014 Apr;29(2):265. doi: 10.1007/s12291-014-0422-8. PMID: 23543587 Free PMC article. Retracted.
-
Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase.Oncotarget. 2012 May;3(5):546-58. doi: 10.18632/oncotarget.503. Oncotarget. 2012. PMID: 22643842 Free PMC article.
-
Optimization of routine KRAS mutation PCR-based testing procedure for rational individualized first-line-targeted therapy selection in metastatic colorectal cancer.Cancer Med. 2013 Feb;2(1):11-20. doi: 10.1002/cam4.47. Epub 2013 Feb 3. Cancer Med. 2013. PMID: 24133623 Free PMC article.
-
KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study.BMJ Open. 2014 May 23;4(5):e004652. doi: 10.1136/bmjopen-2013-004652. BMJ Open. 2014. PMID: 24859998 Free PMC article.
-
High-resolution melting (HRM) assay for the detection of recurrent BRCA1/BRCA2 germline mutations in Tunisian breast/ovarian cancer families.Fam Cancer. 2014 Dec;13(4):603-9. doi: 10.1007/s10689-014-9740-5. Fam Cancer. 2014. PMID: 25069718
References
-
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

