The major histocompatibility complex (Mhc) class IIB region has greater genomic structural flexibility and diversity in the quail than the chicken
- PMID: 17184537
- PMCID: PMC1769493
- DOI: 10.1186/1471-2164-7-322
The major histocompatibility complex (Mhc) class IIB region has greater genomic structural flexibility and diversity in the quail than the chicken
Abstract
Background: The quail and chicken major histocompatibility complex (Mhc) genomic regions have a similar overall organization but differ markedly in that the quail has an expanded number of duplicated class I, class IIB, natural killer (NK)-receptor-like, lectin-like and BG genes. Therefore, the elucidation of genetic factors that contribute to the greater Mhc diversity in the quail would help to establish it as a model experimental animal in the investigation of avian Mhc associated diseases. AIMS AND APPROACHES: The main aim here was to characterize the genetic and genomic features of the transcribed major quail MhcIIB (CojaIIB) region that is located between the Tapasin and BRD2 genes, and to compare our findings to the available information for the chicken MhcIIB (BLB). We used four approaches in the study of the quail MhcIIB region, (1) haplotype analyses with polymorphic loci, (2) cloning and sequencing of the RT-PCR CojaIIB products from individuals with different haplotypes, (3) genomic sequencing of the CojaIIB region from the individuals with the different haplotypes, and (4) phylogenetic and duplication analysis to explain the variability of the region between the quail and the chicken.
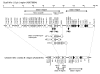
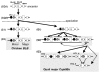
Results: Our results show that the Tapasin-BRD2 segment of the quail Mhc is highly variable in length and in gene transcription intensity and content. Haplotypic sequences were found to vary in length between 4 to 11 kb. Tapasin-BRD2 segments contain one or two major transcribed CojaIIBs that were probably generated by segmental duplications involving c-type lectin-like genes and NK receptor-like genes, gene fusions between two CojaIIBs and transpositions between the major and minor CojaIIB segments. The relative evolutionary speed for generating the MhcIIBs genomic structures from the ancestral BLB2 was estimated to be two times faster in the quail than in the chicken after their separation from a common ancestor. Four types of genomic rearrangement elements (GRE), composed of simple tandem repeats (STR), were identified in the MhcIIB genomic segment located between the Tapasin-BRD2 genes. The GREs have many more STR numbers in the quail than in the chicken that displays strong linkage disequilibrium.
Conclusion: This study suggests that the Mhc classIIB region has a flexible genomic structure generated by rearrangement elements and rapid SNP accumulation probably as a consequence of the quail adapting to environmental conditions and pathogens during its migratory history after its divergence from the chicken.
Figures


Similar articles
-
Comparative genomic analysis of two avian (quail and chicken) MHC regions.J Immunol. 2004 Jun 1;172(11):6751-63. doi: 10.4049/jimmunol.172.11.6751. J Immunol. 2004. PMID: 15153492
-
Sequencing of the core MHC region of black grouse (Tetrao tetrix) and comparative genomics of the galliform MHC.BMC Genomics. 2012 Oct 15;13:553. doi: 10.1186/1471-2164-13-553. BMC Genomics. 2012. PMID: 23066932 Free PMC article.
-
Organisation and diversity of the class II DM region of the chicken MHC.Mol Immunol. 2011 May;48(9-10):1263-71. doi: 10.1016/j.molimm.2011.03.009. Epub 2011 Apr 8. Mol Immunol. 2011. PMID: 21481938
-
Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man.Immunol Rev. 2002 Dec;190:95-122. doi: 10.1034/j.1600-065x.2002.19008.x. Immunol Rev. 2002. PMID: 12493009 Review.
-
Distinctive polymorphism of chicken B-FI (major histocompatibility complex class I) molecules.Poult Sci. 2004 Apr;83(4):600-5. doi: 10.1093/ps/83.4.600. Poult Sci. 2004. PMID: 15109057 Review.
Cited by
-
Long-Read Genome Assemblies Reveal Extraordinary Variation in the Number and Structure of MHC Loci in Birds.Genome Biol Evol. 2021 Feb 3;13(2):evaa270. doi: 10.1093/gbe/evaa270. Genome Biol Evol. 2021. PMID: 33367721 Free PMC article.
-
Trans-species polymorphism of the Mhc class II DRB-like gene in banded penguins (genus Spheniscus).Immunogenetics. 2009 May;61(5):341-52. doi: 10.1007/s00251-009-0363-1. Epub 2009 Mar 25. Immunogenetics. 2009. PMID: 19319519
-
Polymorphism of MHC class IIB in an acheilognathid species, Rhodeus sinensis shaped by historical selection and recombination.BMC Genet. 2019 Sep 13;20(1):74. doi: 10.1186/s12863-019-0775-3. BMC Genet. 2019. PMID: 31519169 Free PMC article.
-
Comparative genome analyses reveal distinct structure in the saltwater crocodile MHC.PLoS One. 2014 Dec 11;9(12):e114631. doi: 10.1371/journal.pone.0114631. eCollection 2014. PLoS One. 2014. PMID: 25503521 Free PMC article.
-
Genetic variation of the major histocompatibility complex (MHC class II B gene) in the threatened Hume's pheasant, Syrmaticus humiae.PLoS One. 2015 Jan 28;10(1):e0116499. doi: 10.1371/journal.pone.0116499. eCollection 2015. PLoS One. 2015. PMID: 25629763 Free PMC article.
References
-
- Klein J. Antigen-major histocompatibility complex-T cell receptors: inquiries into the immunological menage a trois. Immunol Res. 1986;5:173–190. - PubMed
-
- Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, Kaufman J, Zoorob R, Briles WE. 2004 Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics. 2004;56:261–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

