Multiple independent evolutionary solutions to core histone gene regulation
- PMID: 17184543
- PMCID: PMC1794435
- DOI: 10.1186/gb-2006-7-12-r122
Multiple independent evolutionary solutions to core histone gene regulation
Abstract
Background: Core histone genes are periodically expressed along the cell cycle and peak during S phase. Core histone gene expression is deeply evolutionarily conserved from the yeast Saccharomyces cerevisiae to human.
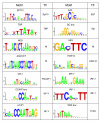
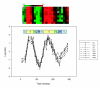
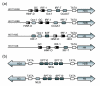
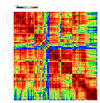
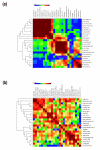
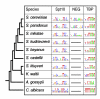
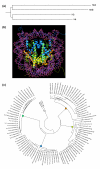
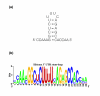
Results: We evaluated the evolutionary dynamics of the specific regulatory mechanisms that give rise to the conserved histone regulatory phenotype. In contrast to the conservation of core histone gene expression patterns, the core histone regulatory machinery is highly divergent between species. There has been substantial evolutionary turnover of cis-regulatory sequence motifs along with the transcription factors that bind them. The regulatory mechanisms employed by members of the four core histone families are more similar within species than within gene families. The presence of species-specific histone regulatory mechanisms is opposite to what is seen at the protein sequence level. Core histone proteins are more similar within families, irrespective of their species of origin, than between families, which is consistent with the shared common ancestry of the members of individual histone families. Structure and sequence comparisons between histone families reveal that H2A and H2B form one related group whereas H3 and H4 form a distinct group, which is consistent with the nucleosome assembly dynamics.
Conclusion: The dissonance between the evolutionary conservation of the core histone gene regulatory phenotypes and the divergence of their regulatory mechanisms indicates a highly dynamic mode of regulatory evolution. This distinct mode of regulatory evolution is probably facilitated by a solution space for promoter sequences, in terms of functionally viable cis-regulatory sites, that is substantially greater than that of protein sequences.
Figures

Similar articles
-
The organization structure and regulatory elements of Chlamydomonas histone genes reveal features linking plant and animal genes.Curr Genet. 1995 Sep;28(4):333-45. doi: 10.1007/BF00326431. Curr Genet. 1995. PMID: 8590479
-
Genomic organization, nucleotide sequence analysis of the core histone genes cluster in Chlamys farreri and molecular evolution assessment of the H2A and H2B.DNA Seq. 2006 Dec;17(6):440-51. doi: 10.1080/10425170600752593. DNA Seq. 2006. PMID: 17381045
-
Nucleotide sequences of Caenorhabditis elegans core histone genes. Genes for different histone classes share common flanking sequence elements.J Mol Biol. 1989 Apr 20;206(4):567-77. doi: 10.1016/0022-2836(89)90566-4. J Mol Biol. 1989. PMID: 2544730
-
Structural comparisons reveal diverse binding modes between nucleosome assembly proteins and histones.Epigenetics Chromatin. 2022 May 24;15(1):20. doi: 10.1186/s13072-022-00452-9. Epigenetics Chromatin. 2022. PMID: 35606827 Free PMC article. Review.
-
Regulation of histone gene expression in budding yeast.Genetics. 2012 May;191(1):7-20. doi: 10.1534/genetics.112.140145. Genetics. 2012. PMID: 22555441 Free PMC article. Review.
Cited by
-
Leveraging Gene Redundancy to Find New Histone Drivers in Cancer.Cancers (Basel). 2023 Jun 30;15(13):3437. doi: 10.3390/cancers15133437. Cancers (Basel). 2023. PMID: 37444547 Free PMC article.
-
Towards a mechanism for histone chaperones.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):211-221. doi: 10.1016/j.bbagrm.2011.07.007. Biochim Biophys Acta. 2013. PMID: 24459723 Free PMC article. Review.
-
Placental transcriptome analysis in opioid-exposed versus non-opioid exposed pregnancies.Placenta. 2025 Mar 25;162:27-34. doi: 10.1016/j.placenta.2025.02.012. Epub 2025 Feb 17. Placenta. 2025. PMID: 39983471
-
Arabidopsis thaliana: a powerful model organism to explore histone modifications and their upstream regulations.Epigenetics. 2023 Dec;18(1):2211362. doi: 10.1080/15592294.2023.2211362. Epigenetics. 2023. PMID: 37196184 Free PMC article. Review.
-
The Evolution of Epigenetics: From Prokaryotes to Humans and Its Biological Consequences.Genet Epigenet. 2016 Aug 3;8:25-36. doi: 10.4137/GEG.S31863. eCollection 2016. Genet Epigenet. 2016. PMID: 27512339 Free PMC article. Review.
References
-
- van Holde KE. Chromatin. London: Springer-Verlag; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

