Evolutionary conservation of domain-domain interactions
- PMID: 17184549
- PMCID: PMC1794438
- DOI: 10.1186/gb-2006-7-12-r125
Evolutionary conservation of domain-domain interactions
Abstract
Background: Recently, there has been much interest in relating domain-domain interactions (DDIs) to protein-protein interactions (PPIs) and vice versa, in an attempt to understand the molecular basis of PPIs.
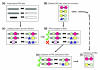
Results: Here we map structurally derived DDIs onto the cellular PPI networks of different organisms and demonstrate that there is a catalog of domain pairs that is used to mediate various interactions in the cell. We show that these DDIs occur frequently in protein complexes and that homotypic interactions (of a domain with itself) are abundant. A comparison of the repertoires of DDIs in the networks of Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens shows that many DDIs are evolutionarily conserved.
Conclusion: Our results indicate that different organisms use the same 'building blocks' for PPIs, suggesting that the functionality of many domain pairs in mediating protein interactions is maintained in evolution.
Figures



Similar articles
-
Systems biology approach reveals possible evolutionarily conserved moonlighting functions for enolase.Comput Biol Chem. 2015 Oct;58:1-8. doi: 10.1016/j.compbiolchem.2015.04.010. Epub 2015 Apr 21. Comput Biol Chem. 2015. PMID: 25978602
-
Protein-protein interactions more conserved within species than across species.PLoS Comput Biol. 2006 Jul 21;2(7):e79. doi: 10.1371/journal.pcbi.0020079. Epub 2006 May 18. PLoS Comput Biol. 2006. PMID: 16854211 Free PMC article.
-
Evolution of multicellularity in Metazoa: comparative analysis of the subcellular localization of proteins in Saccharomyces, Drosophila and Caenorhabditis.Cell Biol Int. 2004;28(3):171-8. doi: 10.1016/j.cellbi.2003.11.016. Cell Biol Int. 2004. PMID: 14984742
-
Inferring protein-protein interactions from multiple protein domain combinations.Methods Mol Biol. 2009;541:43-59. doi: 10.1007/978-1-59745-243-4_3. Methods Mol Biol. 2009. PMID: 19381530 Review.
-
Evaluation of different domain-based methods in protein interaction prediction.Biochem Biophys Res Commun. 2009 Dec 18;390(3):357-62. doi: 10.1016/j.bbrc.2009.09.130. Epub 2009 Oct 2. Biochem Biophys Res Commun. 2009. PMID: 19800868 Review.
Cited by
-
Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria.Microbiol Mol Biol Rev. 2012 Mar;76(1):66-112. doi: 10.1128/MMBR.05011-11. Microbiol Mol Biol Rev. 2012. PMID: 22390973 Free PMC article. Review.
-
PTIR: Predicted Tomato Interactome Resource.Sci Rep. 2016 Apr 28;6:25047. doi: 10.1038/srep25047. Sci Rep. 2016. PMID: 27121261 Free PMC article.
-
In silico Prediction, Characterization, Molecular Docking, and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides Against Fusarium Wilt in Tomato.Front Pharmacol. 2018 Oct 22;9:1038. doi: 10.3389/fphar.2018.01038. eCollection 2018. Front Pharmacol. 2018. PMID: 30405403 Free PMC article.
-
Triangle network motifs predict complexes by complementing high-error interactomes with structural information.BMC Bioinformatics. 2009 Jun 27;10:196. doi: 10.1186/1471-2105-10-196. BMC Bioinformatics. 2009. PMID: 19558694 Free PMC article.
-
Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome.PLoS Biol. 2008 Nov 11;6(11):e277. doi: 10.1371/journal.pbio.0060277. PLoS Biol. 2008. PMID: 18998772 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

