The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA
- PMID: 17185417
- PMCID: PMC1765426
- DOI: 10.1073/pnas.0607063104
The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA
Erratum in
-
Correction for Guo et al., The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA.Proc Natl Acad Sci U S A. 2022 Aug 2;119(31):e2211135119. doi: 10.1073/pnas.2211135119. Epub 2022 Jul 25. Proc Natl Acad Sci U S A. 2022. PMID: 35878045 Free PMC article. No abstract available.
Abstract
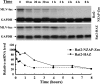
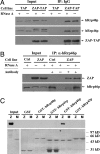
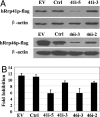
Zinc-finger antiviral protein (ZAP) is a host antiviral factor that specifically inhibits the replication of Moloney murine leukemia virus (MLV) and Sindbis virus (SIN) by preventing accumulation of the viral mRNA in the cytoplasm. In previous studies, we demonstrated that ZAP directly binds to its specific target mRNAs. In this article, we provide evidence indicating that ZAP recruits the RNA processing exosome to degrade the target RNA. ZAP comigrated with the exosome in sucrose or glycerol velocity gradient centrifugation. Immunoprecipitation of ZAP coprecipitated the exosome components. In vitro pull-down assays indicated that ZAP directly interacted with the exosome component hRrp46p and that the binding region of ZAP was mapped to amino acids 224-254. Depletion of the exosome component hRrp41p or hRrp46p with small interfering RNA significantly reduced ZAP's destabilizing activity. These findings suggest that ZAP is a trans-acting factor that modulates mRNA stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. J Cell Physiol. 2003;195:356–372. - PubMed
-
- Guhaniyogi J, Brewer G. Gene. 2001;265:11–23. - PubMed
-
- Khodursky AB, Bernstein JA. Trends Genet. 2003;19:113–115. - PubMed
-
- Vasudevan S, Peltz SW. Curr Opin Cell Biol. 2003;15:332–337. - PubMed
-
- Vasudevan S, Peltz SW, Wilusz CJ. BioEssays. 2002;24:785–788. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

