Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy
- PMID: 17185422
- PMCID: PMC1765419
- DOI: 10.1073/pnas.0604353104
Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy
Abstract
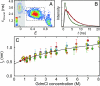
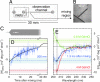
We have used the combination of single-molecule Förster resonance energy transfer and kinetic synchrotron radiation circular dichroism experiments to probe the conformational ensemble of the collapsed unfolded state of the small cold shock protein CspTm under near-native conditions. This regime is physiologically most relevant but difficult to access experimentally, because the equilibrium signal in ensemble experiments is dominated by folded molecules. Here, we avoid this problem in two ways. One is the use of single-molecule Förster resonance energy transfer, which allows the separation of folded and unfolded subpopulations at equilibrium and provides information on long-range intramolecular distance distributions. From experiments with donor and acceptor chromophores placed at different positions within the chain, we find that the distance distributions in unfolded CspTm agree surprisingly well with a Gaussian chain not only at high concentrations of denaturant, where the polypeptide chain is expanded, but also at low denaturant concentrations, where the chain is collapsed. The second, complementary approach is synchrotron radiation circular dichroism spectroscopy of collapsed unfolded molecules transiently populated with a microfluidic device that enables rapid mixing. The results indicate a beta-structure content of the collapsed unfolded state of approximately 20% compared with the folded protein. This suggests that collapse can induce secondary structure in an unfolded state without interfering with long-range distance distributions characteristic of a random coil, which were previously found only for highly expanded unfolded proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
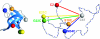


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

