Herpes simplex virus eliminates host mitochondrial DNA
- PMID: 17186027
- PMCID: PMC1796774
- DOI: 10.1038/sj.embor.7400878
Herpes simplex virus eliminates host mitochondrial DNA
Abstract
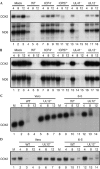
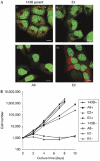
Mitochondria have crucial roles in the life and death of mammalian cells, and help to orchestrate host antiviral defences. Here, we show that the ubiquitous human pathogen herpes simplex virus (HSV) induces rapid and complete degradation of host mitochondrial DNA during productive infection of cultured mammalian cells. The depletion of mitochondrial DNA requires the viral UL12 gene, which encodes a conserved nuclease with orthologues in all herpesviruses. We show that an amino-terminally truncated UL12 isoform-UL12.5-localizes to mitochondria and triggers mitochondrial DNA depletion in the absence of other HSV gene products. By contrast, full-length UL12, a nuclear protein, has little or no effect on mitochondrial DNA levels. Our data document that HSV inflicts massive genetic damage to a crucial host organelle and show a novel mechanism of virus-induced shutoff of host functions, which is likely to contribute to the cell death and tissue damage caused by this widespread human pathogen.
Figures


References
-
- Ashley N, Harris D, Poulton J (2005) Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp Cell Res 303: 432–446 - PubMed
-
- Beal MF (2005) Mitochondria take center stage in aging and neurodegeneration. Ann Neurol 58: 495–505 - PubMed
-
- Chandrasekaran K, Mehrabian Z, Li XL, Hassel B (2004) RNase-L regulates the stability of mitochondrial DNA-encoded mRNAs in mouse embryo fibroblasts. Biochem Biophys Res Commun 325: 18–23 - PubMed
-
- Dimauro S (2004) Mitochondrial medicine. Biochim Biophys Acta 1659: 107–114 - PubMed
-
- Fernandez-Silva P, Enriquez JA, Montoya J (2003) Replication and transcription of mammalian mitochondrial DNA. Exp Physiol 88: 41–56 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

