High performance computing in biology: multimillion atom simulations of nanoscale systems
- PMID: 17187988
- PMCID: PMC1868470
- DOI: 10.1016/j.jsb.2006.10.023
High performance computing in biology: multimillion atom simulations of nanoscale systems
Abstract
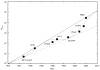
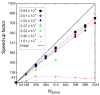
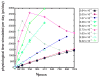
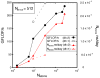
Computational methods have been used in biology for sequence analysis (bioinformatics), all-atom simulation (molecular dynamics and quantum calculations), and more recently for modeling biological networks (systems biology). Of these three techniques, all-atom simulation is currently the most computationally demanding, in terms of compute load, communication speed, and memory load. Breakthroughs in electrostatic force calculation and dynamic load balancing have enabled molecular dynamics simulations of large biomolecular complexes. Here, we report simulation results for the ribosome, using approximately 2.64 million atoms, the largest all-atom biomolecular simulation published to date. Several other nano-scale systems with different numbers of atoms were studied to measure the performance of the NAMD molecular dynamics simulation program on the Los Alamos National Laboratory Q Machine. We demonstrate that multimillion atom systems represent a 'sweet spot' for the NAMD code on large supercomputers. NAMD displays an unprecedented 85% parallel scaling efficiency for the ribosome system on 1024 CPUs. We also review recent targeted molecular dynamics simulations of the ribosome that prove useful for studying conformational changes of this large biomolecular complex in atomic detail.
Figures




Similar articles
-
Scalable Molecular Dynamics with NAMD on the Summit System.IBM J Res Dev. 2018 Nov-Dec;62(6):1-9. doi: 10.1147/jrd.2018.2888986. Epub 2018 Dec 21. IBM J Res Dev. 2018. PMID: 32154805 Free PMC article.
-
Scaling of Multimillion-Atom Biological Molecular Dynamics Simulation on a Petascale Supercomputer.J Chem Theory Comput. 2009 Oct 13;5(10):2798-808. doi: 10.1021/ct900292r. J Chem Theory Comput. 2009. PMID: 26631792
-
Coarse-grained models to study dynamics of nanoscale biomolecules and their applications to the ribosome.J Phys Condens Matter. 2010 Nov 17;22(45):453101. doi: 10.1088/0953-8984/22/45/453101. Epub 2010 Oct 28. J Phys Condens Matter. 2010. PMID: 21339588 Review.
-
Proceedings of the Second Workshop on Theory meets Industry (Erwin-Schrödinger-Institute (ESI), Vienna, Austria, 12-14 June 2007).J Phys Condens Matter. 2008 Feb 13;20(6):060301. doi: 10.1088/0953-8984/20/06/060301. Epub 2008 Jan 24. J Phys Condens Matter. 2008. PMID: 21693862
-
Molecular dynamics simulations of large macromolecular complexes.Curr Opin Struct Biol. 2015 Apr;31:64-74. doi: 10.1016/j.sbi.2015.03.007. Epub 2015 Apr 4. Curr Opin Struct Biol. 2015. PMID: 25845770 Free PMC article. Review.
Cited by
-
Path of nascent polypeptide in exit tunnel revealed by molecular dynamics simulation of ribosome.Biophys J. 2008 Dec 15;95(12):5962-73. doi: 10.1529/biophysj.108.134890. Epub 2008 Oct 20. Biophys J. 2008. PMID: 18936244 Free PMC article.
-
Four-scale description of membrane sculpting by BAR domains.Biophys J. 2008 Sep 15;95(6):2806-21. doi: 10.1529/biophysj.108.132563. Epub 2008 May 30. Biophys J. 2008. PMID: 18515394 Free PMC article.
-
Reconstruction and visualization of large-scale volumetric models of neocortical circuits for physically-plausible in silico optical studies.BMC Bioinformatics. 2017 Sep 13;18(Suppl 10):402. doi: 10.1186/s12859-017-1788-4. BMC Bioinformatics. 2017. PMID: 28929974 Free PMC article.
-
Particle-based methods for multiscale modeling of blood flow in the circulation and in devices: challenges and future directions. Sixth International Bio-Fluid Mechanics Symposium and Workshop March 28-30, 2008 Pasadena, California.Ann Biomed Eng. 2010 Mar;38(3):1225-35. doi: 10.1007/s10439-010-9904-x. Ann Biomed Eng. 2010. PMID: 20336827 Free PMC article.
-
Molecular recognition in the case of flexible targets.Curr Pharm Des. 2011;17(17):1663-71. doi: 10.2174/138161211796355056. Curr Pharm Des. 2011. PMID: 21619526 Free PMC article.
References
-
- Auffinger P, Westhof E. Simulations of the molecular dynamics of nucleic acids. Current Opinion in Structural Biology. 1998;8(2):227–236. - PubMed
-
- Auffinger P, Westhof E. Water and ion binding around r(UpA)(12) and d(TpA)(12) oligomers: Comparison with RNA and DNA (CpG)(12) duplexes. Journal of Molecular Biology. 2001;305(5):1057–1072. - PubMed
-
- Auffinger P, Westhof E. Melting of the solvent structure around a RNA duplex: a molecular dynamics simulation study. Biophys Chem. 2002;95(3):203–210. - PubMed
-
- Board J, Causey J, Leathrum J, Windemuth A, Schulten K. Accelerated molecular dynamics simulation with the parallel fast multipole algorithm. Chemical Physics Letters. 1992;198:89–94.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

