Sequestosome 1/p62--more than just a scaffold
- PMID: 17188686
- PMCID: PMC1850379
- DOI: 10.1016/j.febslet.2006.12.027
Sequestosome 1/p62--more than just a scaffold
Abstract
The interaction of proteins with ubiquitin receptors is key to solving the mystery that surrounds the functional role ubiquitin chains play in directing traffic. The specificity of these interactions is largely mediated by UbL/UBA domains. Sequestosome 1/p62 is a protein that is gaining attention as it is intimately involved in cell signaling, receptor internalization, and protein turnover. Herein we review recent advances in the field.
Figures

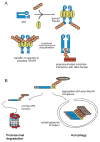

References
-
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. - PubMed
-
- Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. - PubMed
-
- Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

