The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint
- PMID: 17189191
- PMCID: PMC1850967
- DOI: 10.1016/j.molcel.2006.11.027
The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint
Abstract
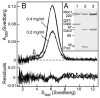
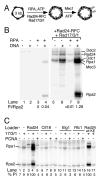
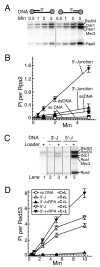
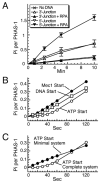
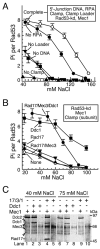
Yeast Mec1/Ddc2 protein kinase, the ortholog of human ATR/ATRIP, plays a central role in the DNA damage checkpoint. The PCNA-like clamp Rad17/Mec3/Ddc1 (the 9-1-1 complex in human) and its loader Rad24-RFC are also essential components of this signal transduction pathway. Here we have studied the role of the clamp in regulating Mec1, and we delineate how the signal generated by DNA lesions is transduced to the Rad53 effector kinase. The checkpoint clamp greatly activates the kinase activity of Mec1, but only if the clamp is appropriately loaded upon partial duplex DNA. Activated Mec1 phosphorylates the Ddc1 and Mec3 subunits of the clamp, the Rad24 subunit of the loader, and the Rpa1 and Rpa2 subunits of RPA. Phosphorylation of Rad53, and of human PHAS-1, a nonspecific target, also requires a properly loaded clamp. Phosphorylation and binding studies with individual clamp subunits indicate that the Ddc1 subunit mediates the functional interactions with Mec1.
Figures

Similar articles
-
Function of Rad17/Mec3/Ddc1 and its partial complexes in the DNA damage checkpoint.DNA Repair (Amst). 2005 Sep 28;4(10):1189-94. doi: 10.1016/j.dnarep.2005.07.008. DNA Repair (Amst). 2005. PMID: 16137930
-
The structure of the checkpoint clamp 9-1-1 complex and clamp loader Rad24-RFC in Saccharomyces cerevisiae.Biochem Biophys Res Commun. 2019 Aug 6;515(4):688-692. doi: 10.1016/j.bbrc.2019.05.138. Epub 2019 Jun 8. Biochem Biophys Res Commun. 2019. PMID: 31182279
-
Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-independent mechanism.PLoS Genet. 2014 Feb 20;10(2):e1004136. doi: 10.1371/journal.pgen.1004136. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24586187 Free PMC article.
-
Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase.Biochem Soc Trans. 2011 Apr;39(2):600-5. doi: 10.1042/BST0390600. Biochem Soc Trans. 2011. PMID: 21428947 Review.
-
A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1.DNA Repair (Amst). 2009 Sep 2;8(9):996-1003. doi: 10.1016/j.dnarep.2009.03.011. Epub 2009 May 22. DNA Repair (Amst). 2009. PMID: 19464966 Free PMC article. Review.
Cited by
-
Assembly of Slx4 signaling complexes behind DNA replication forks.EMBO J. 2015 Aug 13;34(16):2182-97. doi: 10.15252/embj.201591190. Epub 2015 Jun 25. EMBO J. 2015. PMID: 26113155 Free PMC article.
-
Phosphorylation-dependent interactions between Crb2 and Chk1 are essential for DNA damage checkpoint.PLoS Genet. 2012 Jul;8(7):e1002817. doi: 10.1371/journal.pgen.1002817. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792081 Free PMC article.
-
Chromatin remodeling factors Isw2 and Ino80 regulate checkpoint activity and chromatin structure in S phase.Genetics. 2015 Apr;199(4):1077-91. doi: 10.1534/genetics.115.174730. Epub 2015 Feb 19. Genetics. 2015. PMID: 25701287 Free PMC article.
-
The Ddc1-Mec3-Rad17 sliding clamp regulates histone-histone chaperone interactions and DNA replication-coupled nucleosome assembly in budding yeast.J Biol Chem. 2014 Apr 11;289(15):10518-10529. doi: 10.1074/jbc.M114.552463. Epub 2014 Feb 25. J Biol Chem. 2014. PMID: 24573675 Free PMC article.
-
Activation of Tel1ATM kinase requires Rad50 ATPase and long nucleosome-free DNA but no DNA ends.J Biol Chem. 2019 Jun 28;294(26):10120-10130. doi: 10.1074/jbc.RA119.008410. Epub 2019 May 9. J Biol Chem. 2019. PMID: 31073030 Free PMC article.
References
-
- Bartrand AJ, Iyasu D, Brush GS. DNA stimulates Mec1-mediated phosphorylation of replication protein A. J Biol Chem. 2004;279:26762–26767. - PubMed
-
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

