Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice
- PMID: 17189208
- PMCID: PMC3080013
- DOI: 10.1016/j.cmet.2006.11.012
Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice
Abstract
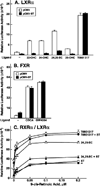
Liver X receptors (LXRs) are nuclear receptors that play crucial roles in lipid metabolism in vivo and are activated by oxysterol ligands in vitro. The identity of the ligand that activates LXRs in vivo is uncertain. Here we provide two lines of evidence that oxysterols are LXR ligands in vitro and in vivo. First, overexpression of an oxysterol catabolic enzyme, cholesterol sulfotransferase, inactivates LXR signaling in several cultured mammalian cell lines but does not alter receptor response to the nonsterol agonist T0901317. Adenovirus-mediated expression of the enzyme in mice prevents dietary induction of hepatic LXR target genes by cholesterol but not by T0901317. Second, triple-knockout mice deficient in the biosynthesis of three oxysterol ligands of LXRs, 24S-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol, respond to dietary T0901317 by inducing LXR target genes in liver but show impaired responses to dietary cholesterol. We conclude that oxysterols are in vivo ligands for LXR.
Figures



References
-
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

