Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation
- PMID: 17189288
- PMCID: PMC3706933
- DOI: 10.1093/hmg/ddl469
Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation
Abstract
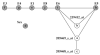
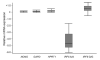
Results from two studies have implicated the interferon regulatory gene IRF5 as a susceptibility gene in systemic lupus erythematosus (SLE). In this study, we conducted a family-based association analysis in 380 UK SLE nuclear families. Using a higher density of markers than has hitherto been screened, we show that there is association with two SNPs in the first intron, rs2004640 (P = 3.4 x 10(-4)) and rs3807306 (P = 4.9 x 10(-4)), and the association extends into the 3'-untranslated region (UTR). There is a single haplotype block encompassing IRF5 and we show for the first time that the gene comprises two over-transmitted haplotypes and a single under-transmitted haplotype. The strongest association is with a TCTAACT haplotype (T:U = 1.92, P = 5.8 x 10(-5)), which carries all the over-transmitted alleles from this study. Haplotypes carrying the T alleles of rs2004640 and rs2280714 and the A allele of rs10954213 are over-transmitted in SLE families. The TAT haplotype shows a dose-dependent relationship with mRNA expression. A differential expression pattern was seen between two expression probes located each side of rs10954213 in the 3'-UTR. rs10954213 shows the strongest association with RNA expression levels (P = 1 x 10(-14)). The A allele of rs10954213 creates a functional polyadenylation site and the A genotype correlates with increased expression of a transcript variant containing a shorter 3'-UTR. Expression levels of transcript variants with the shorter or longer 3'-UTRs are inversely correlated. Our data support a new mechanism by which an IRF5 polymorphism controls the expression of alternate transcript variants which may have different effects on interferon signalling.
Figures



References
-
- Rus V, Hochberg MC. In: Dubois’ Lupus Erythematosus. Wallace DJ, Hahn BH, editors. Lippincott, Williams and Wilkins; Baltimore: 2002. pp. 65–83.
-
- Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–318. - PubMed
-
- Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM. A revised estimate of twin concordance in systemic lupus erythematosus. Arth. Rheum. 1992;35:311–318. - PubMed
-
- Gaffney PM, Ortmann WA, Selby SA, Shark KB, Ockenden TC, Rohlf KE, Walgrave NL, Boyum WP, Malmgren ML, Miller ME, et al. Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analyses of 187 sib-pair families. Am. J. Hum. Genet. 2000;66:547–556. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

