Transcriptional profiling of the Arabidopsis embryo
- PMID: 17189330
- PMCID: PMC1803724
- DOI: 10.1104/pp.106.087668
Transcriptional profiling of the Arabidopsis embryo
Erratum in
- Plant Physiol. 2007 Apr;143(4):1982
Abstract
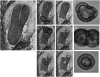
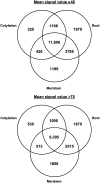
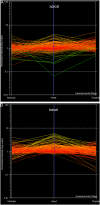
We have used laser-capture microdissection to isolate RNA from discrete tissues of globular, heart, and torpedo stage embryos of Arabidopsis (Arabidopsis thaliana). This was amplified and analyzed by DNA microarray using the Affymetrix ATH1 GeneChip, representing approximately 22,800 Arabidopsis genes. Cluster analysis showed that spatial differences in gene expression were less significant than temporal differences. Time course analysis reveals the dynamics and complexity of gene expression in both apical and basal domains of the developing embryo, with several classes of synexpressed genes identifiable. The transition from globular to heart stage is associated in particular with an up-regulation of genes involved in cell cycle control, transcriptional regulation, and energetics and metabolism. The transition from heart to torpedo stage is associated with a repression of cell cycle genes and an up-regulation of genes encoding storage proteins, and pathways of cell growth, energy, and metabolism. The torpedo stage embryo shows strong functional differentiation in the root and cotyledon, as inferred from the classes of genes expressed in these tissues. The time course of expression of the essential EMBRYO-DEFECTIVE genes shows that most are expressed at unchanging levels across all stages of embryogenesis. We show how identified genes can be used to generate cell type-specific markers and promoter activities for future application in cell biology.
Figures






References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Barton KM, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289–300
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

