Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway
- PMID: 17189340
- PMCID: PMC1785412
- DOI: 10.1105/tpc.106.042473
Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway
Abstract
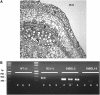
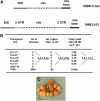
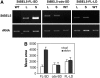
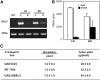
BEL1-like transcription factors interact with Knotted1 types to regulate numerous developmental processes. In potato (Solanum tuberosum), the BEL1 transcription factor St BEL5 and its protein partner POTH1 regulate tuber formation by mediating hormone levels in the stolon tip. The accumulation of St BEL5 RNA increases in response to short-day photoperiods, inductive for tuber formation. RNA detection methods and heterografting experiments demonstrate that BEL5 transcripts are present in phloem cells and move across a graft union to localize in stolon tips, the site of tuber induction. This movement of RNA to stolon tips is correlated with enhanced tuber production. Overexpression of BEL5 transcripts that include the untranslated sequences of the BEL5 transcript endows transgenic lines with the capacity to overcome the inhibitory effects of long days on tuber formation. Addition of the untranslated regions leads to preferential accumulation of the BEL5 RNA in stolon tips under short-day conditions. Using a leaf-specific promoter, the movement of BEL5 RNA to stolon tips was facilitated by a short-day photoperiod, and this movement was correlated with enhanced tuber production. These results implicate the transcripts of St BEL5 in a long-distance signaling pathway that are delivered to the target organ via the phloem stream.
Figures




References
-
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11 36–42.
-
- An, G., Ebert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–19.
-
- Arroyo-Helguera, O., Mejia-Viggiano, C., Varela-Echavarria, A., Cajero-Juarez, M., and Aceves, C. (2005). Regulatory role of the 3′ untranslated region (3′UTR) of rat 5′ deiodinase (D1). Effects on messenger RNA translation and stability. Endocrine 27 219–225. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

