Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms
- PMID: 17189375
- PMCID: PMC1855740
- DOI: 10.1128/JB.01369-06
Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms
Abstract
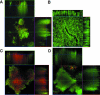
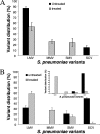
In this study, we report the isolation of colony morphology variants from Streptococcus pneumoniae serotype 3 biofilms. The colony variants differed in colony size (large, medium, and small) and their mucoid appearance on blood agar. The small nonmucoid variant (SCV) emerged during the initial attachment stage of S. pneumoniae biofilm formation and dominated over the course of biofilm growth. Mucoid variants appeared at later biofilm developmental stages. The reduction in colony size/mucoidy correlated with a decrease in capsule production and an increase in initial attachment. The large mucoid variant formed flat unstructured biofilms, failed to aggregate in liquid culture, and adhered poorly to solid surfaces. In contrast, SCVs autoaggregated in liquid culture, hyperadhered to solid surfaces, and formed biofilms with significant three-dimensional structure, mainly in the form of microcolonies. The variants showed similar antibiotic resistance/susceptibility based on a modified Kirby-Bauer test and when grown as biofilms. However, antimicrobial treatment of S. pneumoniae biofilms altered the colony variant's distribution and mainly affected the most interior areas of biofilm microcolonies. To further explore the nature of the variants, the capsule biosynthetic operon (cps3DSUM) was explored in greater detail. The genetic analysis indicated that the emergence of nonmucoid variants was due to a deletion comprising cps3DSU as well as additional genes upstream of the cps3 operon. Overall, our findings suggest that in vitro biofilm formation of S. pneumoniae serotype 3 coincides with the emergence of colony variants with distinct genotypic and phenotypic characteristics.
Figures
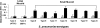


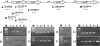

References
-
- Austrian, R. 1981. Some observations on the pneumococcus and the current status of pneumococcal disease and its prevention. Rev. Infect. Dis. 3:S1-S17. - PubMed
-
- Bauer, A. W., D. M. Perry, and W. M. M. Kirby. 1959. Single disc antibiotic sensitivity testing of staphylococci. Arch. Int. Med. 104:208-216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

