Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus
- PMID: 17189377
- PMCID: PMC1855745
- DOI: 10.1128/JB.01658-06
Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus
Abstract
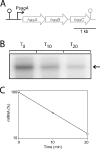
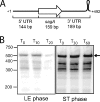
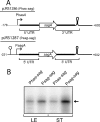
The impressive disease spectrum of Streptococcus pyogenes (the group A streptococcus [GAS]) is believed to be determined by its ability to modify gene expression in response to environmental stimuli. Virulence gene expression is controlled tightly by several different transcriptional regulators in this organism. In addition, expression of most, if not all, GAS genes is determined by a global mechanism dependent on growth phase. To begin an analysis of growth-phase regulation, we compared the transcriptome 2 h into stationary phase to that in late exponential phase of a serotype M3 GAS strain. We identified the arc transcript as more abundant in stationary phase in addition to the sag and sda transcripts that had been previously identified. We found that in stationary phase, the stability of sagA, sda, and arcT transcripts increased dramatically. We found that polynucleotide phosphorylase (PNPase [encoded by pnpA]) is rate limiting for decay of sagA and sda transcripts in late exponential phase, since the stability of these mRNAs was greater in a pnpA mutant, while stability of control mRNAs was unaffected by this mutation. Complementation restored the wild-type decay rate. Furthermore, in a pnpA mutant, the sagA mRNA appeared to be full length, as determined by Northern hybridization. It seems likely that mRNAs abundant in stationary phase are insensitive to the normal decay enzyme(s) and instead require PNPase for this process. It is possible that PNPase activity is limited in stationary phase, allowing persistence of these important virulence factor transcripts at this phase of growth.
Figures


References
-
- Baga, M., M. Goransson, S. Normark, and B. E. Uhlin. 1988. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell 52:197-206. - PubMed
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

