Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans
- PMID: 17189485
- PMCID: PMC1797948
- DOI: 10.1128/EC.00297-06
Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans
Figures


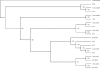
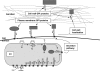
References
-
- Alberti-Segui, C., A. J. Morales, H. Xing, M. M. Kessler, D. A. Willins, K. G. Weinstock, G. Cottarel, K. Fechtel, and B. Rogers. 2004. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 21:285-302. - PubMed
-
- Albrecht, A., A. Felk, I. Pichova, J. R. Naglik, M. Schaller, P. de Groot, D. Maccallum, F. C. Odds, W. Schafer, F. Klis, M. Monod, and B. Hube. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688-694. - PubMed
-
- Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335-1351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

