UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke
- PMID: 17190599
- PMCID: PMC1866287
- DOI: 10.1016/j.cell.2006.10.049
UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke
Abstract
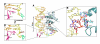
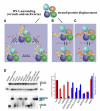
Helicases use the energy derived from nucleoside triphosphate hydrolysis to unwind double helices in essentially every metabolic pathway involving nucleic acids. Earlier crystal structures have suggested that DNA helicases translocate along a single-stranded DNA in an inchworm fashion. We report here a series of crystal structures of the UvrD helicase complexed with DNA and ATP hydrolysis intermediates. These structures reveal that ATP binding alone leads to unwinding of 1 base pair by directional rotation and translation of the DNA duplex, and ADP and Pi release leads to translocation of the developing single strand. Thus DNA unwinding is achieved by a two-part power stroke in a combined wrench-and-inchworm mechanism. The rotational angle and translational distance of DNA define the unwinding step to be 1 base pair per ATP hydrolyzed. Finally, a gateway for ssDNA translocation and an alternative strand-displacement mode may explain the varying step sizes reported previously.
Figures




References
-
- Abdel-Monem M, Chanal MC, Hoffmann-Berling H. DNA unwinding enzyme II of Escherichia coli. 1. Purification and characterization of the ATPase activity. Eur J Biochem. 1977;79:33–38. - PubMed
-
- Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. - PubMed
-
- Arthur HM, Lloyd RG. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. - PubMed
-
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

