A critical role for the autophagy gene Atg5 in T cell survival and proliferation
- PMID: 17190837
- PMCID: PMC2118420
- DOI: 10.1084/jem.20061303
A critical role for the autophagy gene Atg5 in T cell survival and proliferation
Abstract
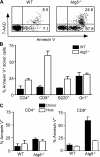
Macroautophagy (hereafter referred to as autophagy) is a well-conserved intracellular degradation process. Recent studies examining cells lacking the autophagy genes Atg5 and Atg7 have demonstrated that autophagy plays essential roles in cell survival during starvation, in innate cell clearance of microbial pathogens, and in neural cell maintenance. However, the role of autophagy in T lymphocyte development and survival is not known. Here, we demonstrate that autophagosomes form in primary mouse T lymphocytes. By generating Atg5-/- chimeric mice, we found that Atg5-deficient T lymphocytes underwent full maturation. However, the numbers of total thymocytes and peripheral T and B lymphocytes were reduced in Atg5 chimeras. In the periphery, Atg5-/- CD8+ T lymphocytes displayed dramatically increased cell death. Furthermore, Atg5-/- CD4+ and CD8+ T cells failed to undergo efficient proliferation after TCR stimulation. These results demonstrate a critical role for Atg5 in multiple aspects of lymphocyte development and function and suggest that autophagy may be essential for both T lymphocyte survival and proliferation.
Figures



References
-
- Mizushima, N., Y. Ohsumi, and T. Yoshimori. 2002. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27:421–429. - PubMed
-
- Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. - PubMed
-
- Mizushima, N. 2005. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12(Suppl 2):1535–1541. - PubMed
-
- Lum, J.J., R.J. DeBerardinis, and C.B. Thompson. 2005. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6:439–448. - PubMed
-
- Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

