Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array
- PMID: 17192196
- PMCID: PMC1769375
- DOI: 10.1186/1471-2164-7-325
Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array
Abstract
Background: Alternative splicing is a mechanism for increasing protein diversity by excluding or including exons during post-transcriptional processing. Alternatively spliced proteins are particularly relevant in oncology since they may contribute to the etiology of cancer, provide selective drug targets, or serve as a marker set for cancer diagnosis. While conventional identification of splice variants generally targets individual genes, we present here a new exon-centric array (GeneChip Human Exon 1.0 ST) that allows genome-wide identification of differential splice variation, and concurrently provides a flexible and inclusive analysis of gene expression.
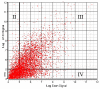
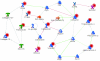
Results: We analyzed 20 paired tumor-normal colon cancer samples using a microarray designed to detect over one million putative exons that can be virtually assembled into potential gene-level transcripts according to various levels of prior supporting evidence. Analysis of high confidence (empirically supported) transcripts identified 160 differentially expressed genes, with 42 genes occupying a network impacting cell proliferation and another twenty nine genes with unknown functions. A more speculative analysis, including transcripts based solely on computational prediction, produced another 160 differentially expressed genes, three-fourths of which have no previous annotation. We also present a comparison of gene signal estimations from the Exon 1.0 ST and the U133 Plus 2.0 arrays. Novel splicing events were predicted by experimental algorithms that compare the relative contribution of each exon to the cognate transcript intensity in each tissue. The resulting candidate splice variants were validated with RT-PCR. We found nine genes that were differentially spliced between colon tumors and normal colon tissues, several of which have not been previously implicated in cancer. Top scoring candidates from our analysis were also found to substantially overlap with EST-based bioinformatic predictions of alternative splicing in cancer.
Conclusion: Differential expression of high confidence transcripts correlated extremely well with known cancer genes and pathways, suggesting that the more speculative transcripts, largely based solely on computational prediction and mostly with no previous annotation, might be novel targets in colon cancer. Five of the identified splicing events affect mediators of cytoskeletal organization (ACTN1, VCL, CALD1, CTTN, TPM1), two affect extracellular matrix proteins (FN1, COL6A3) and another participates in integrin signaling (SLC3A2). Altogether they form a pattern of colon-cancer specific alterations that may particularly impact cell motility.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

