Linkage disequilibrium of evolutionarily conserved regions in the human genome
- PMID: 17192199
- PMCID: PMC1769491
- DOI: 10.1186/1471-2164-7-326
Linkage disequilibrium of evolutionarily conserved regions in the human genome
Abstract
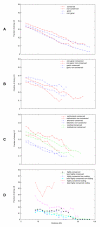
Background: The strong linkage disequilibrium (LD) recently found in genic or exonic regions of the human genome demonstrated that LD can be increased by evolutionary mechanisms that select for functionally important loci. This suggests that LD might be stronger in regions conserved among species than in non-conserved regions, since regions exposed to natural selection tend to be conserved. To assess this hypothesis, we used genome-wide polymorphism data from the HapMap project and investigated LD within DNA sequences conserved between the human and mouse genomes.
Results: Unexpectedly, we observed that LD was significantly weaker in conserved regions than in non-conserved regions. To investigate why, we examined sequence features that may distort the relationship between LD and conserved regions. We found that interspersed repeats, and not other sequence features, were associated with the weak LD tendency in conserved regions. To appropriately understand the relationship between LD and conserved regions, we removed the effect of repetitive elements and found that the high degree of sequence conservation was strongly associated with strong LD in coding regions but not with that in non-coding regions.
Conclusion: Our work demonstrates that the degree of sequence conservation does not simply increase LD as predicted by the hypothesis. Rather, it implies that purifying selection changes the polymorphic patterns of coding sequences but has little influence on the patterns of functional units such as regulatory elements present in non-coding regions, since the former are generally restricted by the constraint of maintaining a functional protein product across multiple exons while the latter may exist more as individually isolated units.
Figures
Similar articles
-
Assessing the patterns of linkage disequilibrium in genic regions of the human genome.FEBS J. 2011 Oct;278(19):3748-55. doi: 10.1111/j.1742-4658.2011.08293.x. Epub 2011 Sep 8. FEBS J. 2011. PMID: 21824289
-
Small fitness effect of mutations in highly conserved non-coding regions.Hum Mol Genet. 2005 Aug 1;14(15):2221-9. doi: 10.1093/hmg/ddi226. Epub 2005 Jun 30. Hum Mol Genet. 2005. PMID: 15994173
-
Increased gene coverage and Alu frequency in large linkage disequilibrium blocks of the human genome.Genet Mol Res. 2007 Dec 11;6(4):1131-41. Genet Mol Res. 2007. PMID: 18273807
-
Raising the estimate of functional human sequences.Genome Res. 2007 Sep;17(9):1245-53. doi: 10.1101/gr.6406307. Epub 2007 Aug 9. Genome Res. 2007. PMID: 17690206 Review.
-
Conserved non-genic sequences - an unexpected feature of mammalian genomes.Nat Rev Genet. 2005 Feb;6(2):151-7. doi: 10.1038/nrg1527. Nat Rev Genet. 2005. PMID: 15716910 Review.
Cited by
-
The heterogeneous levels of linkage disequilibrium in white spruce genes and comparative analysis with other conifers.Heredity (Edinb). 2012 Mar;108(3):273-84. doi: 10.1038/hdy.2011.72. Epub 2011 Sep 7. Heredity (Edinb). 2012. PMID: 21897435 Free PMC article.
-
Investigation of founder effects for the Thr377Met Myocilin mutation in glaucoma families from differing ethnic backgrounds.Mol Vis. 2007 Mar 28;13:487-92. Mol Vis. 2007. PMID: 17417609 Free PMC article.
-
Population-genetic nature of copy number variations in the human genome.Hum Mol Genet. 2010 Mar 1;19(5):761-73. doi: 10.1093/hmg/ddp541. Epub 2009 Dec 5. Hum Mol Genet. 2010. PMID: 19966329 Free PMC article.
References
-
- Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, Carter D, Papaspyridonos M, Livingstone S, Ganske R, Lohmussaar E, Zernant J, Tonisson N, Remm M, Magi R, Puurand T, Vilo J, Kurg A, Rice K, Deloukas P, Mott R, Metspalu A, Bentley DR, Cardon LR, Dunham I. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418:544–548. doi: 10.1038/nature00864. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

