Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins
- PMID: 17192395
- PMCID: PMC6485507
- DOI: 10.1182/blood-2006-07-036269
Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins
Abstract
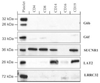
To identify previously unknown platelet receptors we compared the transcriptomes of in vitro differentiated megakaryocytes (MKs) and erythroblasts (EBs). RNA was obtained from purified, biologically paired MK and EB cultures and compared using cDNA microarrays. Bioinformatical analysis of MK-up-regulated genes identified 151 transcripts encoding transmembrane domain-containing proteins. Although many of these were known platelet genes, a number of previously unidentified or poorly characterized transcripts were also detected. Many of these transcripts, including G6b, G6f, LRRC32, LAT2, and the G protein-coupled receptor SUCNR1, encode proteins with structural features or functions that suggest they may be involved in the modulation of platelet function. Immunoblotting on platelets confirmed the presence of the encoded proteins, and flow cytometric analysis confirmed the expression of G6b, G6f, and LRRC32 on the surface of platelets. Through comparative analysis of expression in platelets and other blood cells we demonstrated that G6b, G6f, and LRRC32 are restricted to the platelet lineage, whereas LAT2 and SUCNR1 were also detected in other blood cells. The identification of the succinate receptor SUCNR1 in platelets is of particular interest, because physiologically relevant concentrations of succinate were shown to potentiate the effect of low doses of a variety of platelet agonists.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures

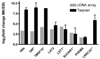
 ) and real-time PCR (
) and real-time PCR ( ) are plotted as Log2 (fold change). Results are shown as the average for the 5-paired MK/EB comparisons. Although all transcripts were detected by RT-PCR in all 5 of the MK samples, transcripts marked with an asterisk (*) were not detected in all of the EB cultures. Thus, the ratio is based on fewer than 5 comparisons (G6f, n = 2; TMSF15, n = 1; LST1 and SUCNR1, n = 4; all others n = 5). **LRRC32 was identified as differentially expressed in preliminary array experiments and by Taqman real-time PCR. Error bars represent SD of replicates.
) are plotted as Log2 (fold change). Results are shown as the average for the 5-paired MK/EB comparisons. Although all transcripts were detected by RT-PCR in all 5 of the MK samples, transcripts marked with an asterisk (*) were not detected in all of the EB cultures. Thus, the ratio is based on fewer than 5 comparisons (G6f, n = 2; TMSF15, n = 1; LST1 and SUCNR1, n = 4; all others n = 5). **LRRC32 was identified as differentially expressed in preliminary array experiments and by Taqman real-time PCR. Error bars represent SD of replicates.


References
-
- McDonald TP, Sullivan PS. Megakaryocytic and erythrocytic cell lines share a common precursor cell. Exp Hematol. 1993;21:1316–1320. - PubMed
-
- Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. - PubMed
-
- Martin DI, Zon LI, Mutter G, Orkin SH. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. - PubMed
-
- Romeo PH, Prandini MH, Joulin V, et al. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344:447–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

