Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization
- PMID: 17192411
- PMCID: PMC1805085
- DOI: 10.1091/mbc.e06-08-0707
Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization
Abstract
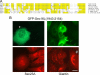
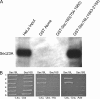
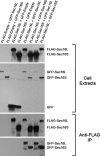
Budding yeast Sec16 is a large peripheral endoplasmic reticulum (ER) membrane protein that functions in generating COPII transport vesicles and in clustering COPII components at transitional ER (tER) sites. Sec16 interacts with multiple COPII components. Although the COPII assembly pathway is evolutionarily conserved, Sec16 homologues have not been described in higher eukaryotes. Here, we show that mammalian cells contain two distinct Sec16 homologues: a large protein that we term Sec16L and a smaller protein that we term Sec16S. These proteins localize to tER sites, and an N-terminal region of each protein is necessary and sufficient for tER localization. The Sec16L and Sec16S genes are both expressed in every tissue examined, and both proteins are required in HeLa cells for ER export and for normal tER organization. Sec16L resembles yeast Sec16 in having a C-terminal conserved domain that interacts with the COPII coat protein Sec23, but Sec16S lacks such a C-terminal conserved domain. Immunoprecipitation data indicate that Sec16L and Sec16S are each present at multiple copies in a heteromeric complex. We infer that mammalian cells have preserved and extended the function of Sec16.
Figures

References
-
- Bevis B. J., Hammond A. T., Reinke C. A., Glick B. S. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 2002;4:750–756. - PubMed
-
- Connerly P. L., Esaki M., Montegna E. A., Strongin D. E., Levi S., Soderholm J., Glick B. S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005;15:1439–1447. - PubMed
-
- Duden R., Schekman R. Insights into Golgi function through mutants in yeast and animal cells. In: Berger E. G., Roth J., editors. The Golgi Apparatus. Basel, Switzerland: Birkhäuser Verlag; 1997. pp. 219–246.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

