The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer
- PMID: 17192413
- PMCID: PMC1805106
- DOI: 10.1091/mbc.e06-06-0542
The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer
Abstract
Cullins are members of a family of scaffold proteins that assemble multisubunit ubiquitin ligase complexes to confer substrate specificity for the ubiquitination pathway. Cullin3 (Cul3) forms a catalytically inactive BTB-Cul3-Rbx1 (BCR) ubiquitin ligase, which becomes functional upon covalent attachment of the ubiquitin homologue neural-precursor-cell-expressed and developmentally down regulated 8 (Nedd8) near the C terminus of Cul3. Current models suggest that Nedd8 activates cullin complexes by providing a recognition site for a ubiquitin-conjugating enzyme. Based on the following evidence, we propose that Nedd8 activates the BCR ubiquitin ligase by mediating the dimerization of Cul3. First, Cul3 is found as a neddylated heterodimer bound to a BTB domain-containing protein in vivo. Second, the formation of a Cul3 heterodimer is mediated by a Nedd8 molecule, which covalently attaches itself to one Cul3 molecule and binds to the winged-helix B domain at the C terminus of the second Cul3 molecule. Third, complementation experiments revealed that coexpression of two distinct nonfunctional Cul3 mutants can rescue the ubiquitin ligase function of the BCR complex. Likewise, a substrate of the BCR complex binds heterodimeric Cul3, suggesting that the Cul3 complex is active as a dimer. These findings not only provide insight into the architecture of the active BCR complex but also suggest assembly as a regulatory mechanism for activation of all cullin-based ubiquitin ligases.
Figures





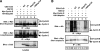

References
-
- Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 2006;8:348–357. - PubMed
-
- Clurman B. E., Sheaff R. J., Thress K., Groudine M., Roberts J. M. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. - PubMed
-
- Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. - PubMed
-
- Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. BTB/POZ domain proteins are putative substrate adaptors for Cullin 3 ubiquitin ligases. Mol. Cell. 2003;12:783–790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

