Upregulation of the neuron-specific K+/Cl- cotransporter expression by transcription factor early growth response 4
- PMID: 17192429
- PMCID: PMC6674722
- DOI: 10.1523/JNEUROSCI.4731-06.2006
Upregulation of the neuron-specific K+/Cl- cotransporter expression by transcription factor early growth response 4
Abstract
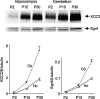
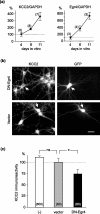
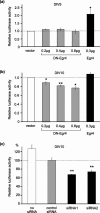
The expression of the neuron-specific K+/Cl- cotransporter (KCC2) is restricted to the CNS and is strongly upregulated during neuronal maturation, yielding a low intracellular chloride concentration that is required for fast synaptic inhibition in adult neurons. To elucidate the mechanisms of KCC2 gene regulation, we analyzed the KCC2 (alias Slc12a5) promoter and proximal intron-1 regions and revealed 10 candidate transcription factor binding sites that are highly conserved in mammalian KCC2 genes. Here we focus on one of these factors, early growth response 4 (Egr4), which shows a similar developmental upregulation in CNS neurons as KCC2. KCC2 luciferase reporter constructs containing the Egr4 site (Egr4(KCC2)) were strongly induced by Egr4 overexpression in neuro-2a neuroblastoma cells and in cultured neurons. Egr4-mediated induction was decreased significantly by point-mutating the Egr4(KCC2). Insertion of Egr4(KCC2) into the KCC2 basal promoter in the endogenous reverse, but not in the opposite, orientation reestablished Egr4-mediated induction. Electrophoretic mobility shift assay confirmed specific Egr4 binding to Egr4(KCC2). Interference RNA-mediated knock-down of Egr4 and a dominant-negative isoform of Egr4 significantly inhibited KCC2 reporter induction and endogenous KCC2 expression in cultured neurons. Together, the results indicate an important role for Egr4 in the developmental upregulation of KCC2 gene expression.
Figures





Similar articles
-
Early growth response 4 mediates BDNF induction of potassium chloride cotransporter 2 transcription.J Neurosci. 2011 Jan 12;31(2):644-9. doi: 10.1523/JNEUROSCI.2006-10.2011. J Neurosci. 2011. PMID: 21228173 Free PMC article.
-
Neurturin evokes MAPK-dependent upregulation of Egr4 and KCC2 in developing neurons.Neural Plast. 2011;2011:1-8. doi: 10.1155/2011/641248. Epub 2011 Jul 4. Neural Plast. 2011. PMID: 21837281 Free PMC article.
-
Neuronal K+/Cl- co-transporter (KCC2) transgenes lacking neurone restrictive silencer element recapitulate CNS neurone-specific expression and developmental up-regulation of endogenous KCC2 gene.J Neurochem. 2005 Nov;95(4):1144-55. doi: 10.1111/j.1471-4159.2005.03434.x. J Neurochem. 2005. PMID: 16271048
-
Crossing the Chloride Channel: The Current and Potential Therapeutic Value of the Neuronal K+-Cl- Cotransporter KCC2.Biomed Res Int. 2019 May 21;2019:8941046. doi: 10.1155/2019/8941046. eCollection 2019. Biomed Res Int. 2019. PMID: 31240228 Free PMC article. Review.
-
The K(+)-Cl(-) Cotransporter KCC2 and Chloride Homeostasis: Potential Therapeutic Target in Acute Central Nervous System Injury.Mol Neurobiol. 2016 May;53(4):2141-51. doi: 10.1007/s12035-015-9162-x. Epub 2015 May 5. Mol Neurobiol. 2016. PMID: 25941074 Review.
Cited by
-
Transcriptome analysis of mouse brain infected with Toxoplasma gondii.Infect Immun. 2013 Oct;81(10):3609-19. doi: 10.1128/IAI.00439-13. Epub 2013 Jul 15. Infect Immun. 2013. PMID: 23856619 Free PMC article.
-
The primate-specific peptide Y-P30 regulates morphological maturation of neocortical dendritic spines.PLoS One. 2019 Feb 13;14(2):e0211151. doi: 10.1371/journal.pone.0211151. eCollection 2019. PLoS One. 2019. PMID: 30759095 Free PMC article.
-
The DNA repair protein ATM as a target in autism spectrum disorder.JCI Insight. 2021 Feb 8;6(3):e133654. doi: 10.1172/jci.insight.133654. JCI Insight. 2021. PMID: 33373327 Free PMC article.
-
Early growth response 4 mediates BDNF induction of potassium chloride cotransporter 2 transcription.J Neurosci. 2011 Jan 12;31(2):644-9. doi: 10.1523/JNEUROSCI.2006-10.2011. J Neurosci. 2011. PMID: 21228173 Free PMC article.
-
Reciprocal Regulation of KCC2 Trafficking and Synaptic Activity.Front Cell Neurosci. 2019 Feb 20;13:48. doi: 10.3389/fncel.2019.00048. eCollection 2019. Front Cell Neurosci. 2019. PMID: 30842727 Free PMC article. Review.
References
-
- Aguado F, Carmona MA, Pozas E, Aguilo A, Martinez-Guijarro FJ, Alcantara S, Borrell V, Yuste R, Ibanez CF, Soriano E. BDNF regulates spontaneous correlated activity at early developmental stages by increasing synaptogenesis and expression of the K+/Cl− co-transporter KCC2. Development. 2003;130:1267–1280. - PubMed
-
- Banker G, Goslin K. Ed 2. Cambridge, MA: MIT; 1998. Culturing nerve cells.
-
- Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int. 1997;31:477–510. - PubMed
-
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
