A phosphate-binding subsite in bovine pancreatic ribonuclease A can be converted into a very efficient catalytic site
- PMID: 17192592
- PMCID: PMC2222832
- DOI: 10.1110/ps.062251707
A phosphate-binding subsite in bovine pancreatic ribonuclease A can be converted into a very efficient catalytic site
Abstract
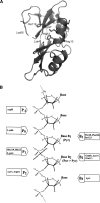
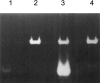
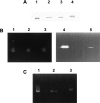
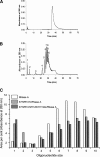
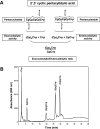
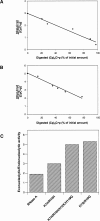
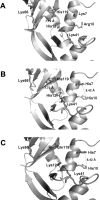
A general acid-base catalytic mechanism is responsible for the cleavage of the phosphodiester bonds of the RNA by ribonuclease A (RNase A). The main active site is formed by the amino acid residues His12, His119, and Lys41, and the process follows an endonucleolytic pattern that depends on the existence of a noncatalytic phosphate-binding subsite adjacent, on the 3'-side, to the active site; in this region the phosphate group of the substrate establishes electrostatic interactions through the side chains of Lys7 and Arg10. We have obtained, by means of site-directed mutagenesis, RNase A variants with His residues both at positions 7 and 10. These mutations have been introduced with the aim of transforming a noncatalytic binding subsite into a putative new catalytic active site. The RNase activity of these variants was determined by the zymogram technique and steady-state kinetic parameters were obtained by spectrophotometric methods. The variants showed a catalytic efficiency in the same order of magnitude as the wild-type enzyme. However, we have demonstrated in these variants important effects on the substrate's cleavage pattern. The quadruple mutant K7H/R10H/H12K/H119Q shows a clear increase of the exonucleolytic activity; in this case the original native active site has been suppressed, and, as consequence, its activity can only be associated to the new active site. In addition, the mutant K7H/R10H, with two putative active sites, also shows an increase in the exonucleolytic preference with respect to the wild type, a fact that may be correlated with the contribution of the new active site.
Figures
References
-
- Alonso, J., Nogués, M.V., and Cuchillo, C.M. 1986. Modification of bovine pancreatic ribonuclease A with 6-chloropurine riboside. Arch. Biochem. Biophys. 246: 681–689. - PubMed
-
- Boix, E., Nogués, M.V., Schein, C.H., Benner, S.A., and Cuchillo, C.M. 1994. Reverse transphosphorylation by ribonuclease A needs an intact p2-binding site. Point mutations at Lys-7 and Arg-10 alter the catalytic properties of the enzyme. J. Biol. Chem. 269: 2529–2534. - PubMed
-
- Boix, E., Leonidas, D.D., Nikolovski, Z., Nogués, M.V., Cuchillo, C.M., and Acharya, K.R. 1999a. Crystal structure of eosinophil cationic protein at 2.4 Å resolution. Biochemistry 38: 16794–16801. - PubMed
-
- Boix, E., Nikolovski, Z., Moiseyev, G.P., Rosenberg, H.F., Cuchillo, C.M., and Nogués, M.V. 1999b. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a site arrangement that favors exonuclease-type activity. J. Biol. Chem. 274: 15605–15614. - PubMed
-
- Bravo, J., Fernández, E., Ribó, M., de Llorens, R., and Cuchillo, C.M. 1994. A versatile negative-staining ribonuclease zymogram. Anal. Biochem. 219: 82–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

