Tissue-specific differences in human transfer RNA expression
- PMID: 17194224
- PMCID: PMC1713254
- DOI: 10.1371/journal.pgen.0020221
Tissue-specific differences in human transfer RNA expression
Abstract
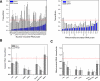
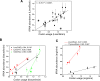
Over 450 transfer RNA (tRNA) genes have been annotated in the human genome. Reliable quantitation of tRNA levels in human samples using microarray methods presents a technical challenge. We have developed a microarray method to quantify tRNAs based on a fluorescent dye-labeling technique. The first-generation tRNA microarray consists of 42 probes for nuclear encoded tRNAs and 21 probes for mitochondrial encoded tRNAs. These probes cover tRNAs for all 20 amino acids and 11 isoacceptor families. Using this array, we report that the amounts of tRNA within the total cellular RNA vary widely among eight different human tissues. The brain expresses higher overall levels of nuclear encoded tRNAs than every tissue examined but one and higher levels of mitochondrial encoded tRNAs than every tissue examined. We found tissue-specific differences in the expression of individual tRNA species, and tRNAs decoding amino acids with similar chemical properties exhibited coordinated expression in distinct tissue types. Relative tRNA abundance exhibits a statistically significant correlation to the codon usage of a collection of highly expressed, tissue-specific genes in a subset of tissues or tRNA isoacceptors. Our findings demonstrate the existence of tissue-specific expression of tRNA species that strongly implicates a role for tRNA heterogeneity in regulating translation and possibly additional processes in vertebrate organisms.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

