Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis
- PMID: 17194304
- PMCID: PMC1769492
- DOI: 10.1186/1471-2164-7-327
Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis
Abstract
Background: Recently, genomic sequencing efforts were finished for Oryza sativa (cultivated rice) and Arabidopsis thaliana (Arabidopsis). Additionally, these two plant species have extensive cDNA and expressed sequence tag (EST) libraries. We employed the Program to Assemble Spliced Alignments (PASA) to identify and analyze alternatively spliced isoforms in both species.
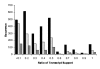
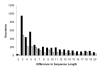
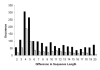
Results: A comprehensive analysis of alternative splicing was performed in rice that started with >1.1 million publicly available spliced ESTs and over 30,000 full length cDNAs in conjunction with the newly enhanced PASA software. A parallel analysis was performed with Arabidopsis to compare and ascertain potential differences between monocots and dicots. Alternative splicing is a widespread phenomenon (observed in greater than 30% of the loci with transcript support) and we have described nine alternative splicing variations. While alternative splicing has the potential to create many RNA isoforms from a single locus, the majority of loci generate only two or three isoforms and transcript support indicates that these isoforms are generally not rare events. For the alternate donor (AD) and acceptor (AA) classes, the distance between the splice sites for the majority of events was found to be less than 50 basepairs (bp). In both species, the most frequent distance between AA is 3 bp, consistent with reports in mammalian systems. Conversely, the most frequent distance between AD is 4 bp in both plant species, as previously observed in mouse. Most alternative splicing variations are localized to the protein coding sequence and are predicted to significantly alter the coding sequence.
Conclusion: Alternative splicing is widespread in both rice and Arabidopsis and these species share many common features. Interestingly, alternative splicing may play a role beyond creating novel combinations of transcripts that expand the proteome. Many isoforms will presumably have negative consequences for protein structure and function, suggesting that their biological role involves post-transcriptional regulation of gene expression.
Figures

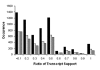
References
-
- Expressed Sequence Tag database dbEST http://www.ncbi.nlm.nih.gov/dbEST
-
- DNA Data Bank of Japan (DDBJ) http://www.ddbj.nig.ac.jp - PubMed
-
- TIGR Rice Genome Annotation Database http://rice.tigr.org
-
- Haas BJ, Delche AL, Mount SM, Wortmann JR, Smith RK, Jr, Hannick LI, Maiti R, Ronning C, Rusch DB, Town CD, Salzberg SL, White O. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

