NMR structure of stem-loop D from human rhinovirus-14
- PMID: 17194719
- PMCID: PMC1800519
- DOI: 10.1261/rna.313707
NMR structure of stem-loop D from human rhinovirus-14
Abstract
The 5'-cloverleaf of the picornavirus RNA genome is essential for the assembly of a ribonucleoprotein replication complex. Stem-loop D (SLD) of the cloverleaf is the recognition site for the multifunctional viral protein 3Cpro. This protein is the principal viral protease, and its interaction with SLD also helps to position the viral RNA-dependent RNA polymerase (3Dpol) for replication. Human rhinovirus-14 (HRV-14) is distinct from the majority of picornaviruses in that its SLD forms a cUAUg triloop instead of the more common uYACGg tetraloop. This difference appears to be functionally significant, as 3Cpro from tetraloop-containing viruses cannot bind the HRV-14 SLD. We have determined the solution structure of the HRV-14 SLD using NMR spectroscopy. The structure is predominantly an A-form helix, but with a central pyrimidine-pyrimidine base-paired region and a significantly widened major groove. The stabilizing hydrogen bonding present in the uYACGg tetraloop was not found in the cUAUg triloop. However, the triloop uses different structural elements to present a largely similar surface: sequence and underlying architecture are not conserved, but key aspects of the surface structure are. Important structural differences do exist, though, and may account for the observed cross-isotype binding specificities between 3Cpro and SLD.
Figures

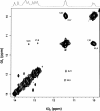

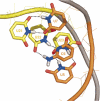
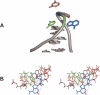
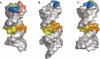

Similar articles
-
The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C.Structure. 2004 Feb;12(2):237-48. doi: 10.1016/j.str.2004.01.014. Structure. 2004. PMID: 14962384
-
Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses.Biochemistry. 2004 Sep 28;43(38):11959-72. doi: 10.1021/bi048973p. Biochemistry. 2004. PMID: 15379536
-
Structure of an RNA hairpin from HRV-14.Biochemistry. 2001 Jul 10;40(27):8055-64. doi: 10.1021/bi010572b. Biochemistry. 2001. PMID: 11434774
-
Human rhinovirus 3C protease as a potential target for the development of antiviral agents.Curr Protein Pept Sci. 2007 Feb;8(1):19-27. doi: 10.2174/138920307779941523. Curr Protein Pept Sci. 2007. PMID: 17305557 Review.
-
RNA structure at high resolution.FASEB J. 1995 Aug;9(11):1023-33. doi: 10.1096/fasebj.9.11.7544309. FASEB J. 1995. PMID: 7544309 Review.
Cited by
-
Role of RNA Domain Structure and Orientation in the Coxsackievirus B3 Virulence Phenotype.J Virol. 2023 May 31;97(5):e0044823. doi: 10.1128/jvi.00448-23. Epub 2023 Apr 19. J Virol. 2023. PMID: 37074194 Free PMC article.
-
Independent evolution of tetraloop in enterovirus oriL replicative element and its putative binding partners in virus protein 3C.PeerJ. 2017 Oct 6;5:e3896. doi: 10.7717/peerj.3896. eCollection 2017. PeerJ. 2017. PMID: 29018627 Free PMC article.
-
Structure of RNA Stem Loop B from the Picornavirus Replication Platform.Biochemistry. 2017 May 23;56(20):2549-2557. doi: 10.1021/acs.biochem.7b00141. Epub 2017 May 5. Biochemistry. 2017. PMID: 28459542 Free PMC article.
-
Exploration of Potential Broad-Spectrum Antiviral Targets in the Enterovirus Replication Element: Identification of Six Distinct 5' Cloverleaves.Viruses. 2024 Jun 23;16(7):1009. doi: 10.3390/v16071009. Viruses. 2024. PMID: 39066172 Free PMC article.
-
Conformational flexibility in the enterovirus RNA replication platform.RNA. 2019 Mar;25(3):376-387. doi: 10.1261/rna.069476.118. Epub 2018 Dec 21. RNA. 2019. PMID: 30578285 Free PMC article.
References
-
- Allain, F.H., Varani, G. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 1995;250:333–353. - PubMed
-
- Amineva, S.P., Aminev, A.G., Palmenberg, A.C., Gern, J.E. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J. Gen. Virol. 2004;85:2969–2979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
