Structure of aspartoacylase, the brain enzyme impaired in Canavan disease
- PMID: 17194761
- PMCID: PMC1766406
- DOI: 10.1073/pnas.0607817104
Structure of aspartoacylase, the brain enzyme impaired in Canavan disease
Abstract
Aspartoacylase catalyzes hydrolysis of N-acetyl-l-aspartate to aspartate and acetate in the vertebrate brain. Deficiency in this activity leads to spongiform degeneration of the white matter of the brain and is the established cause of Canavan disease, a fatal progressive leukodystrophy affecting young children. We present crystal structures of recombinant human and rat aspartoacylase refined to 2.8- and 1.8-A resolution, respectively. The structures revealed that the N-terminal domain of aspartoacylase adopts a protein fold similar to that of zinc-dependent hydrolases related to carboxypeptidases A. The catalytic site of aspartoacylase shows close structural similarity to those of carboxypeptidases despite only 10-13% sequence identity between these proteins. About 100 C-terminal residues of aspartoacylase form a globular domain with a two-stranded beta-sheet linker that wraps around the N-terminal domain. The long channel leading to the active site is formed by the interface of the N- and C-terminal domains. The C-terminal domain is positioned in a way that prevents productive binding of polypeptides in the active site. The structures revealed that residues 158-164 may undergo a conformational change that results in opening and partial closing of the channel entrance. We hypothesize that the catalytic mechanism of aspartoacylase is closely analogous to that of carboxypeptidases. We identify residues involved in zinc coordination, and propose which residues may be involved in substrate binding and catalysis. The structures also provide a structural framework necessary for understanding the deleterious effects of many missense mutations of human aspartoacylase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
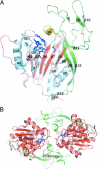


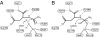
Comment in
-
The impact of structural biology on neurobiology.Proc Natl Acad Sci U S A. 2007 Jan 9;104(2):399-400. doi: 10.1073/pnas.0610164103. Epub 2007 Jan 3. Proc Natl Acad Sci U S A. 2007. PMID: 17213329 Free PMC article. No abstract available.
References
-
- Baslow MH. Neurochem Res. 2003;28:941–953. - PubMed
-
- Clark JF, Doepke A, Filosa JA, Wardle RL, Lu A, Meeker TJ, Pyne-Geithman GJ. Med Hypotheses. 2006;67:506–512. - PubMed
-
- Tsai G, Coyle JT. Prog Neurobiol. 1995;46:531–540. - PubMed
-
- Gujar SK, Maheshwari S, Bjorkman-Burtscher I, Sundgren PC. J Neuroophthalmol. 2005;25:217–226. - PubMed
-
- Madhavarao CN, Chinopoulos C, Chandrasekaran K, Namboodiri MA. J Neurochem. 2003;86:824–835. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

