The carnegie protein trap library: a versatile tool for Drosophila developmental studies
- PMID: 17194782
- PMCID: PMC1840051
- DOI: 10.1534/genetics.106.065961
The carnegie protein trap library: a versatile tool for Drosophila developmental studies
Abstract
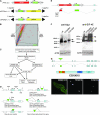
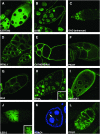
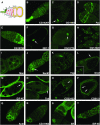
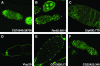
Metazoan physiology depends on intricate patterns of gene expression that remain poorly known. Using transposon mutagenesis in Drosophila, we constructed a library of 7404 protein trap and enhancer trap lines, the Carnegie collection, to facilitate gene expression mapping at single-cell resolution. By sequencing the genomic insertion sites, determining splicing patterns downstream of the enhanced green fluorescent protein (EGFP) exon, and analyzing expression patterns in the ovary and salivary gland, we found that 600-900 different genes are trapped in our collection. A core set of 244 lines trapped different identifiable protein isoforms, while insertions likely to act as GFP-enhancer traps were found in 256 additional genes. At least 8 novel genes were also identified. Our results demonstrate that the Carnegie collection will be useful as a discovery tool in diverse areas of cell and developmental biology and suggest new strategies for greatly increasing the coverage of the Drosophila proteome with protein trap insertions.
Figures
References
-
- Aspenstrom, P., 1997. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7: 479–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

