Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa
- PMID: 17194795
- PMCID: PMC1899365
- DOI: 10.1128/JB.01688-06
Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa
Abstract
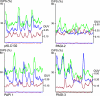
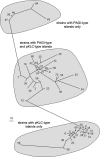
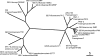
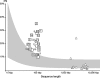
The known genomic islands of Pseudomonas aeruginosa clone C strains are integrated into tRNA(Lys) (pKLC102) or tRNA(Gly) (PAGI-2 and PAGI-3) genes and differ from their core genomes by distinctive tetranucleotide usage patterns. pKLC102 and the related island PAPI-1 from P. aeruginosa PA14 were spontaneously mobilized from their host chromosomes at frequencies of 10% and 0.3%, making pKLC102 the most mobile genomic island known with a copy number of 30 episomal circular pKLC102 molecules per cell. The incidence of islands of the pKLC102/PAGI-2 type was investigated in 71 unrelated P. aeruginosa strains from diverse habitats and geographic origins. pKLC102- and PAGI-2-like islands were identified in 50 and 31 strains, respectively, and 15 and 10 subtypes were differentiated by hybridization on pKLC102 and PAGI-2 macroarrays. The diversity of PAGI-2-type islands was mainly caused by one large block of strain-specific genes, whereas the diversity of pKLC102-type islands was primarily generated by subtype-specific combination of gene cassettes. Chromosomal loss of PAGI-2 could be documented in sequential P. aeruginosa isolates from individuals with cystic fibrosis. PAGI-2 was present in most tested Cupriavidus metallidurans and Cupriavidus campinensis isolates from polluted environments, demonstrating the spread of PAGI-2 across habitats and species barriers. The pKLC102/PAGI-2 family is prevalent in numerous beta- and gammaproteobacteria and is characterized by high asymmetry of the cDNA strands. This evolutionarily ancient family of genomic islands retained its oligonucleotide signature during horizontal spread within and among taxa.
Figures
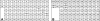


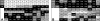


References
-
- Almagor, H. 1983. A Markov analysis of DNA sequences. J. Theor. Biol. 104:633-645. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl, ed. 1994. Current protocols in molecular biology. Wiley, New York, NY.
-
- Baisnee, P. F., S. Hampson, and P. Baldi. 2002. Why are complementary DNA strands symmetric? Bioinformatics 18:1021-1033. - PubMed
-
- Baldi, P., and P. F. Baisnee. 2000. Sequence analysis by additive scales: DNA structure for sequences and repeats of all lengths. Bioinformatics 16:865-889. - PubMed
-
- Bragonzi, A., L. Wiehlmann, J. Klockgether, N. Cramer, D. Worlitzsch, G. Döring, and B. Tümmler. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261-3269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

